
Volume 23, Number 2—February 2017
Dispatch
Persistent Infections with Diverse Co-Circulating Astroviruses in Pediatric Oncology Patients, Memphis, Tennessee, USA
On This Page
Valerie Cortez, Pamela Freiden, Zhengming Gu, Elisabeth Adderson, Randall Hayden, and Stacey Schultz-Cherry
Abstract
Human astroviruses are a major cause of pediatric gastroenteritis, especially in immunocompromised children. We conducted a retrospective study to demonstrate that diverse astrovirus genotypes can co-circulate in pediatric oncology patients. A subset of cases is associated with long-term virus shedding (range 17–183 days).
Astroviruses are a leading cause of diarrhea, and children <2 years of age and immunocompromised persons are at higher risk for systemic and severe disease (1). Few studies have investigated the diversity of astroviruses that infect these populations despite there being 3 distinct phylogenetic clades of human astroviruses (HAstVs) (canonical genotypes HAstV1–8 and noncanonical genotypes MLB1–3 and VA1–5), which makes surveillance challenging (2). To explore the genetic diversity of astroviruses in persons with high-risk for infection, we performed a retrospective study in immunocompromised pediatric oncology patients, analyzing remnant fecal samples collected in 2008 and 2010–2011.
A total of 909 remnant fecal samples were collected from 419 patients; 473 samples from 220 patients were collected during January–December 2008, and 436 samples from 199 patients were collected during January 2010–June 2011 (Technical Appendix[PDF - 512 KB - 6 pages] Figure 1). All samples were de-identified, so that only a patient identification code, sample identification code, and date were known. No other clinical data were available. The St. Jude Children’s Research Hospital Institutional Review Board approved this study with a waiver of consent.
We extracted viral RNA from homogenized fecal samples by using the MagMAX-96 Viral RNA Isolation Kit (Applied Biosystems Life Technologies, Carlsbad, CA, USA). We screened samples from 2008 by using a singleplex real-time reverse transcription PCR (rRT-PCR) (3), an in-house multiplex PCR to identify canonical (HAstV1–8) and noncanonical (MLB1 and VA2) genotypes (4), and endpoint PCRs by using primers targeting open reading frame (ORF) 1(5) and ORF2 (6,7). The singeplex method detected HAstV in 23/72 samples, compared with 61/67 (5 samples were not tested) by the multiplex method. We screened samples from 2010–2011 solely by the multiplex method; 14 were positive for HAstV. Overall, we detected HAstV in 86 (9.5%) of 909 samples from 60 (14.3%) of 419 patients (Table; Technical Appendix[PDF - 512 KB - 6 pages] Table), giving a detection rate comparable to those reported for other immunocompromised populations (8,9).
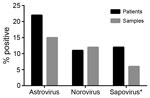
Figure 1. Enteric virus infections identified from remnant fecal samples from pediatric patients with cancer, Memphis, Tennessee, USA, 2008. The percentage of samples and patients testing positive for human astrovirus was higher than...
To place these findings in the context of those for other enteric virus infections, we also used rRT-PCR methods to determine the prevalence of norovirus and sapovirus in the fecal samples from 2008 (10,11). Due to sample quantity limitations, we were unable to test 31 samples for sapovirus. We detected HAstV in the highest proportion of patients and samples (22% [49/220] and 15% [72/473], respectively), followed by norovirus (11% [39/220] and 12% [58/473]) and sapovirus (12% [25/211] and 6.6% [29/442]) (Figure 1). Together, these data demonstrate that HAstVs are a major contributor to the enteric virus infections in this patient population. Furthermore, co-infection occurred in only 16% of HAstV-positive patients (Table; Technical Appendix[PDF - 512 KB - 6 pages] Table), much lower than the 33%–65% of patients reported in other studies (2).
A median of 40 (range 18–54) samples were collected each month in 2008; cases peaked in spring and decreased in summer and early fall (Technical Appendix[PDF - 512 KB - 6 pages] Figure 2). We further investigated temporal trends in HAstV infections by using longitudinal samples from 34 patients (2–12 samples/patient) collected over a median of 81 (range 3–328) days. Of the 34 patients, 12 were previously HAstV negative, indicating that more than one third of patients had newly acquired infections (Technical Appendix[PDF - 512 KB - 6 pages] Table). Of the 12 patients with >1 positive specimen, 6 experienced prolonged HAstV shedding (defined by positive samples collected >2 weeks apart [range 17–183 days]) (Technical Appendix[PDF - 512 KB - 6 pages] Table). Subsequent fecal samples for 2 patients (SJ35 and SJ209) were negative for HAstV, suggesting that the virus had cleared, whereas the other 4 patients (SJ22, SJ175, SJ245, and SJ275) had detectable HAstV in their final fecal samples. Despite being co-infected with HAstV and norovirus, patient SJ275 persistently shed only HAstV, highlighting the need to further explore enteric virus co-infections and the potential for virus interference.
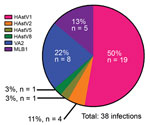
Figure 2. Co-circulating human astrovirus (HAstV) strains in pediatric patients with cancer in Memphis, Tennessee, USA. Six different HAstV genotypes were identified among the HAstV-positive fecal samples collected from pediatric patients in 2008...
We genotyped 50 of the 86 HAstV-positive samples by using the aforementioned endpoint PCR methods, with patient-specific primers and the 3′ RACE System (Invitrogen Life Technologies, Carlsbad, CA, USA) for rapid amplification of cDNA ends to obtain partial open reading frame (ORF) 1b (RNA-dependent RNA polymerase) and ORF2 (capsid protein) sequences. We were unable to genotype the remaining positive samples because of inadequate sample quality or quantity. We used 50 sequences, collectively representing 38 unique infections, in the phylogenetic analysis (Technical Appendix[PDF - 512 KB - 6 pages] Figure 3) and BLAST (https://blast.ncbi.nlm.nih.gov) searches for samples with only shorter sequence reads available. Tree topology of ORF1b and ORF2 sequences indicated that none of the samples contained recombinant viruses or was co-infected with different genotypes. Overall, HAstV1 was the most prevalent genotype identified (n = 19, 50%), followed by the noncanonical genotypes VA2 (n = 8, 21%) and MLB1 (n = 5, 13%) (Figure 2). Fecal samples from 4 (11%) patients were positive for HAstV2; 1 each was positive for HAstV5 and HAstV8. Of note, 4 patients were sequentially infected with >1 genotype, and 3 patients from 2008 had serial infections with canonical and noncanonical genotypes (Table; Technical Appendix[PDF - 512 KB - 6 pages] Table). The average interval between these infections was ≈7 months.
In a previous study of immunocompromised children hospitalized with diarrhea, the prevalence of HAstV infection was 5% (9). HAstV detection in HIV-infected persons of all ages with and without gastroenteritis has been reported to be as high as 12% (8). Thus, the 14% detection rate we observed in pediatric oncology patients is consistent with the rate in previous reports. Some patients showed prolonged virus shedding, which has been reported in immunocompetent and immunocompromised children, in some cases for as long as 3 months (12,13). However, 4 of the patients in our study shed virus beyond 3 months, including 1 who shed for ≈6 months, highlighting the ability of HAstV infections to persist within this population. In addition, 3 of the 4 patients with prolonged shedding were infected with the noncanonical genotypes MLB1 or VA2. Although genotypic differences in virus load and long-term shedding have been reported for canonical viruses (HAstV1–8) (12), we are not aware of such studies with noncanonical viruses. A larger, prospectively followed, longitudinal cohort would be required to investigate these differences.
Our identification of 4 patients with sequential HAstV infections is notable, especially because all cases occurred during 1 year of observation. One previous report also described a child who was first infected with HAstV3 and then, 9 months later, with HAstV1 (14). Although our study showed more than one third of the HAstV infections detected in samples from 2008 were newly acquired, we do not know the precise circumstances of these infections—whether they were associated with healthcare settings, or if they occurred at particular times during the patients’ treatment. The trend of HAstV infections in our study appears to mirror that in reports from Egypt, Brazil, and the eastern United States, which also observed a peak of infections in late spring and early summer (13).
In summary, we used multiple molecular methods to identify HAstV infections in immunocompromised pediatric oncology patients, enabling us to examine a broad representation of the infections experienced by this population and to identify 6 co-circulating viruses. We showed that the singleplex rRT-PCR method (3) was unable to capture infections caused by noncanonical viruses and is, therefore, limited in its utility for accurate surveillance and diagnosis. Although our multiplex method was able to identify all canonical and 2 noncanonical viruses (4), it is possible that we missed infections with other strains. Given the limited knowledge about HAstV genotypes and clinical disease, particularly in immunocompromised persons, additional studies will be crucial to help develop better diagnostics to capture all known HAstV genotypes.
Dr. Cortez is a postdoctoral research fellow at St. Jude Children’s Research Hospital, studying astroviruses and influenza. Her research interests include RNA virus evolution and zoonotic disease transmission.
Acknowledgments
We thank Andrew Burnham, Alexandra Rivera, and Tiara Rainier for technical assistance and David Boyd and Keith Laycock for helpful comments during the preparation of this manuscript.
This study was funded by the Hartwell Foundation, ALSAC (American Lebanese Syrian Associated Charities), a St. Jude Children’s Infection Defense Center grant (to S.S.-C.), and a National Institutes of Health T32 training grant (AI106700 to V.C).
References
- Vu DL, Cordey S, Brito F, Kaiser L. Novel human astroviruses: Novel human diseases? J Clin Virol. 2016;82:56–63. DOIPubMed
- Bosch A, Guix S, Pintó RM. Epidemiology of human astroviruses. In: Schultz-Cherry S, editor. Astrovirus research. New York: Springer; 2012. p. 1–18.
- Podkolzin AT, Fenske EB, Abramycheva NY, Shipulin GA, Sagalova OI, Mazepa VN, et al. Hospital-based surveillance of rotavirus and other viral agents of diarrhea in children and adults in Russia, 2005-2007. J Infect Dis. 2009;200(Suppl 1):S228–33. DOIPubMed
- Gu Z, Zhu H, Rodriguez A, Mhaissen M, Schultz-Cherry S, Adderson E, et al. Comparative evaluation of broad-panel PCR assays for the detection of gastrointestinal pathogens in pediatric oncology patients. J Mol Diagn. 2015;17:715–21. DOIPubMed
- Chu DKW, Poon LLM, Guan Y, Peiris JSM. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–14. DOIPubMed
- Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol. 1995;33:797–801.PubMed
- Finkbeiner SR, Le BM, Holtz LR, Storch GA, Wang D. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg Infect Dis. 2009;15:441–4. DOIPubMed
- Liste MB, Natera I, Suarez JA, Pujol FH, Liprandi F, Ludert JE. Enteric virus infections and diarrhea in healthy and human immunodeficiency virus-infected children. J Clin Microbiol. 2000;38:2873–7.PubMed
- Treviño M, Prieto E, Peñalver D, Aguilera A, García-Zabarte A, García-Riestra C, et al. [Diarrhea caused by adenovirus and astrovirus in hospitalized immunodeficient patients]. Enferm Infecc Microbiol Clin. 2001;19:7–10. DOIPubMed
- Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu F-T, et al. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J Med Virol. 2006;78:1347–53. DOIPubMed
- Stals A, Baert L, Botteldoorn N, Werbrouck H, Herman L, Uyttendaele M, et al. Multiplex real-time RT-PCR for simultaneous detection of GI/GII noroviruses and murine norovirus 1. J Virol Methods. 2009;161:247–53. DOIPubMed
- Caballero S, Guix S, El-Senousy WM, Calicó I, Pintó RM, Bosch A. Persistent gastroenteritis in children infected with astrovirus: association with serotype-3 strains. J Med Virol. 2003;71:245–50. DOIPubMed
- Walter JE, Mitchell DK. Astrovirus infection in children. Curr Opin Infect Dis. 2003;16:247–53. DOIPubMed
- Guix S, Caballero S, Villena C, Bartolomé R, Latorre C, Rabella N, et al. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J Clin Microbiol. 2002;40:133–9. DOIPubMed





















.jpg)












No hay comentarios:
Publicar un comentario