
Best Documentary: Cells Record Their Own Lives Using CRISPR
Suppose you were a police detective investigating a robbery. You’d appreciate having a stack of photographs of the crime in progress, but you’d be even happier if you had a detailed movie of the robbery. Then, you could see what happened and when. Research on cells is somewhat like this. Until recently, scientists could take snapshots of cells in action, but they had trouble recording what cells were doing over time. They were getting an incomplete picture of the events occurring in cells.
Researchers have started solving this problem by combining some old knowledge—that DNA is spectacularly good at storing information—with a popular new research tool called CRISPR. CRISPR (clustered regularly interspaced short palindromic repeats) is an immune system feature in bacteria that helps them to remember and destroy viruses that infect them. CRISPR can change DNA sequences in precise ways; and it’s programmable, meaning that researchers can tell CRISPR where to make a change on a DNA strand, and even what kind of change to make. By linking cellular events to CRISPR, researchers can make something like a movie that captures many pieces of information in the form of DNA changes that researchers can read back later. These pieces of information include timing, duration, and intensity of events, such as the turning on of a specific protein pathway or the exposure of the cell to pathogens (i.e. germs). Here, we look at some of the ways NIGMS-funded research teams and others are using CRISPR to capture these kinds of data within DNA sequences.
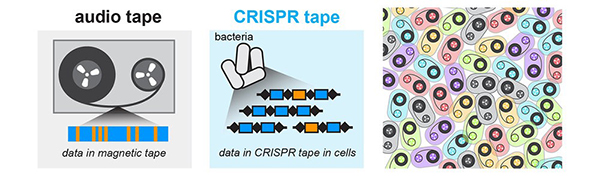
An audio recorder stores audio signals into a magnetic tape medium (left). Similarly, a microscopic data recorder stores biological signals into a CRISPR tape in bacteria (middle). An enormous amount of information can be stored across multiple bacterial cells (right). Credit: Wang Lab/Columbia University Medical Center.
Round and Round: mSCRIBE Creates a Continuous Recording Loop
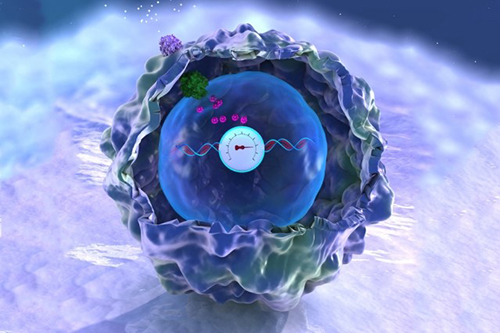
MIT bioengineers, led by Timothy Lu, have devised a memory storage system illustrated here as a DNA-embedded meter that records the activity of a signaling pathway in a human cell. Credit: Timothy Lu lab, MIT.
CRISPR uses an enzyme called Cas9 like a surgical knife, to slice both strands of a cell’s DNA at precise points. A cut like this sends the cell scrambling to repair the damage. Often, the repair effort results in changes, or errors, in the repaired strand that pile up at a known rate. Timothy Lu  and his colleagues at the Massachusetts Institute of Technology (MIT) decided to turn this cut-repair-error system into a way to record certain events inside a cell. They call their method mSCRIBE (mammalian synthetic cellular recorder integrating biological events).
and his colleagues at the Massachusetts Institute of Technology (MIT) decided to turn this cut-repair-error system into a way to record certain events inside a cell. They call their method mSCRIBE (mammalian synthetic cellular recorder integrating biological events).
With mSCRIBE, researchers can link Cas9 activity to an event in the cell they want to know more about. In one test, for example, Cas9 only becomes active when a protein pathway that responds to inflammation turns on. Only when the inflammation pathway turns off will mSCRIBE’s cut-and-repair loop stop. By looking at how long the loop ran, Lu and his team were able to measure how long the inflammation response lasted as well as how intensely it was activated.
Roll ‘Em! CAMERA 1
Instead of letting cells’ repair systems make changes to DNA, as happens in mSCRIBE, David Liu  and Weixin Tang of the Broad Institute of MIT and Harvard coupled CRISPR-Cas9 with other methods that also take advantage of DNA’s ability to act as an event recorder. Their tool called CAMERA 1, adds numerous copies of two plasmids—short strands of DNA—to cells. As in mSCRIBE, Cas9 is linked to an event in the cell. As the event occurs, Cas9 begins destroying copies of one type of plasmid. Researchers can then look at the ratio of the two types of plasmids in the cell to determine how often the event took place.
and Weixin Tang of the Broad Institute of MIT and Harvard coupled CRISPR-Cas9 with other methods that also take advantage of DNA’s ability to act as an event recorder. Their tool called CAMERA 1, adds numerous copies of two plasmids—short strands of DNA—to cells. As in mSCRIBE, Cas9 is linked to an event in the cell. As the event occurs, Cas9 begins destroying copies of one type of plasmid. Researchers can then look at the ratio of the two types of plasmids in the cell to determine how often the event took place.
MEMOIR Uses CRISPR and Imaging Technology to Record Cells’ Histories
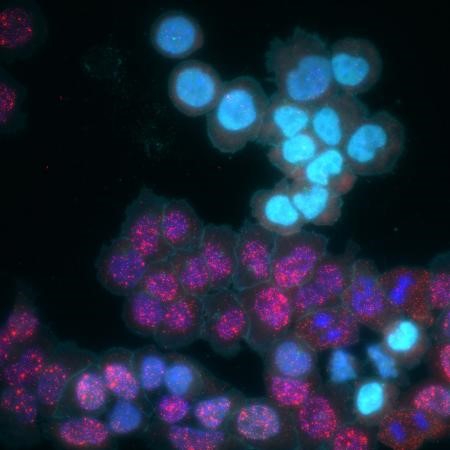
MEMOIR enables cells’ histories to be recorded in their genomes and then read out using microscopy. Here, MEMOIR cells have recorded information in response to a signal with the help of the CRISPR DNA-editing system (light blue cells). Credit: Elowitz and Cai Labs/Caltech.
When studying the changes that cells go through in a living tissue or embryo, researchers ideally want to capture what happens in individual cells without removing them from their surroundings, so their findings more closely relate to real-life events. Many of the CRISPR recording techniques, however, must read the DNA sequence that Cas9 has changed to get their results. This means using at least a few cells and removing them from their tissues to extract the DNA.
Long Cai  and Michael Elowitz
and Michael Elowitz  at the California Institute of Technology in Pasadena blended CRISPR with imaging technology to build a recorder called MEMOIR (memory by engineered mutagenesis with optical in situ readout). MEMOIR allows the team to find and identify CRISPR-caused errors without removing the cell, its tissue, or the larger colony.
at the California Institute of Technology in Pasadena blended CRISPR with imaging technology to build a recorder called MEMOIR (memory by engineered mutagenesis with optical in situ readout). MEMOIR allows the team to find and identify CRISPR-caused errors without removing the cell, its tissue, or the larger colony.
According to Cai, using imaging to read MEMOIR’s results allows all the cells to remain in the tissue or embryo scientists are studying. This is especially important in research on embryos, where small numbers of cells move around to form new tissues and organs during development.
With TRACE, Adding Short Sequences Records Cellular Events
Harris Wang and colleagues at Columbia University Medical Center in New York developed another technique to better track cellular activity. They’re refining a system called TRACE (temporal recording in arrays by CRISPR expansion) to convert bacteria into tiny devices that record what happens when they travel through the human gut, for example. Like Liu’s technique, their system relies on specially designed small loops of DNA. Instead of cutting DNA, however, Wang and his team use CRISPR-Cas to add two different, short sequences to the loop’s genome. The researchers add one sequence—the timekeeper—at a steady rate. They can then monitor the length of the DNA loop. The longer it gets, the more time has passed. The other sequence is tied to an event in the cell. It’s added to the loop only when the event occurs. The resulting strand shows what happened and when, just like a movie.
and colleagues at Columbia University Medical Center in New York developed another technique to better track cellular activity. They’re refining a system called TRACE (temporal recording in arrays by CRISPR expansion) to convert bacteria into tiny devices that record what happens when they travel through the human gut, for example. Like Liu’s technique, their system relies on specially designed small loops of DNA. Instead of cutting DNA, however, Wang and his team use CRISPR-Cas to add two different, short sequences to the loop’s genome. The researchers add one sequence—the timekeeper—at a steady rate. They can then monitor the length of the DNA loop. The longer it gets, the more time has passed. The other sequence is tied to an event in the cell. It’s added to the loop only when the event occurs. The resulting strand shows what happened and when, just like a movie.
Harris Wang
As CRISPR becomes more precise and easier to use the hope, says MIT’s Lu, is that eventually these systems will be able to give scientists a very complex view of what happens inside cells over time. This knowledge can then be useful for fixing cells when they break down in the course of normal aging and disease.
Lu’s research is funded by NIH grants DP2OD008435 and P50GM098792. Liu’s research is funded in part by NIH grants RM1HG009490, R01EB022376, and R35GM118062. Cai’s research is funded in part by NIH grants R01HD075605 and K99GM118910. Wang’s research is funded in part by NIH grant 1DP50D009172.





















.png)
.png)











No hay comentarios:
Publicar un comentario