
Viral Views: New Insights on Infection Strategies
Cutting-edge research tools are giving us clearer views of viruses—and possible ways to disarm them.
Viral Views: New Insights on Infection Strategies
The following images show a few ways in which cutting-edge research tools are giving us clearer views of viruses—and possible ways to disarm them. The examples, which highlight work involving HIV and the coronavirus, were funded in part by our Biomedical Technology Research Resources program.
Uncloaking HIV’s Camouflage
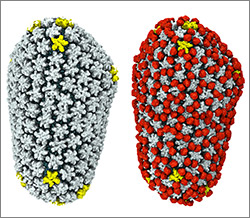
HIV capsid with (right, red) and without (left) a camouflaging human protein. Credit: Juan R. Perilla, Klaus Schulten and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign.
To sneak past our immune defenses and infect human cells, HIV uses a time-honored strategy—disguise. The virus’ genome is enclosed in a protein shell called a capsid (on left) that’s easily recognized and destroyed by the human immune system. To evade this fate, the chrysalis-shaped capsid cloaks itself with a human protein known as cyclophilin A (in red, on right). Camouflaged as human, the virus gains safe passage into and through a human cell to deposit its genetic material in the nucleus and start taking control of cellular machinery.
Biomedical and technical experts teamed up to generate these HIV models at near-atomic resolution. First, structural biologists at the Pittsburgh Center for HIV Protein Interactions  used a technique called cryo-electron microscopy (cryo-EM) to get information on the shape of an HIV capsid as well as the capsid-forming proteins’ connections to each other and to cyclophilin A. Then experts at the Resource for Macromolecular Modeling and Bioinformatics
used a technique called cryo-electron microscopy (cryo-EM) to get information on the shape of an HIV capsid as well as the capsid-forming proteins’ connections to each other and to cyclophilin A. Then experts at the Resource for Macromolecular Modeling and Bioinformatics  fed the cryo-EM data into their visualization and simulation programs to computationally model the physical interactions among every single atom of the capsid and the cyclophilin A protein. The work revealed a previously unknown site where cyclophilin A binds to the capsid, offering new insights on the biology of HIV infection.
fed the cryo-EM data into their visualization and simulation programs to computationally model the physical interactions among every single atom of the capsid and the cyclophilin A protein. The work revealed a previously unknown site where cyclophilin A binds to the capsid, offering new insights on the biology of HIV infection.
The work was published in Nature Communications and was funded in part by NIH under grantsR01GM085043, P50GM082251, P41GM104601, R01GM067887 and P30GM110758.
Decrowning the Coronavirus
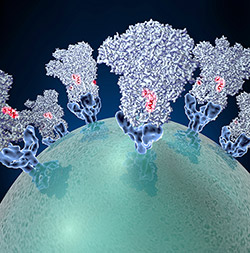
Coronavirus (green globe) with glycoproteins (blue cone-shaped structures) studding the surface. Credit: David Veesler, University of Washington.
To enter cells, the coronavirus uses a common infection strategy—fusion. Like the crests of a crown, glycoproteins (blue cone-shaped structures) stud the virus’ surface (green globe). These proteins attach to proteins on the surface of host cells and cause the virus and cell to fuse together. This fusion allows the virus to dump its genetic material into the host, initiating the infection process. Different strains of the coronavirus can cause mild, cold-like symptoms. In recent years, new strains have emerged that cause deadly respiratory syndromes transmitted from other animals to humans.
The lack of good structural data has prevented scientists from conducting detailed studies of the virus’ fusion machinery—a detailed model may be helpful in developing vaccines and antiviral drugs to prevent infection. To get a sharper picture of the virus’ glycoproteins, scientists at the National Resource for Automated Molecular Microscopy  used cryo-EM to capture structural information about a coronavirus that infects mice. The scientists applied their state-of-the-art methods and tools for collecting and processing cryo-EM data to generate an accurate model of the glyocoproteins. The new model revealed that the outer edge of the protein has a chain of amino acids (a peptide) that’s involved in viral entry and likely a feature of other coronaviruses. Because the peptide is easily accessible, it could be a prime target for potential virus-neutralizing therapies.
used cryo-EM to capture structural information about a coronavirus that infects mice. The scientists applied their state-of-the-art methods and tools for collecting and processing cryo-EM data to generate an accurate model of the glyocoproteins. The new model revealed that the outer edge of the protein has a chain of amino acids (a peptide) that’s involved in viral entry and likely a feature of other coronaviruses. Because the peptide is easily accessible, it could be a prime target for potential virus-neutralizing therapies.
The work was published in Nature and was funded in part by NIH under grants P41GM103310 andT32GM008268.
High-resolution versions of these virus structures and other scientific images, illustrations and videos are freely available in the NIGMS Image and Video Gallery.


































No hay comentarios:
Publicar un comentario