Types of Influenza Viruses
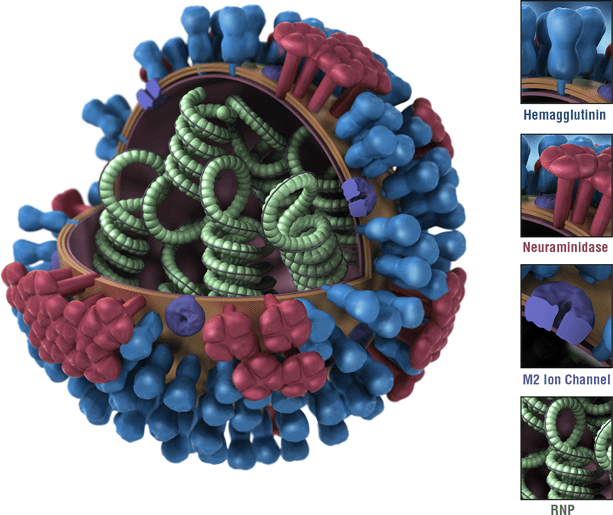
This is a picture of an influenza virus. Influenza A viruses are classified by subtypes based on the properties of their hemagglutinin (H) and neuraminidase (N) surface proteins. There are 18 different HA subtypes and 11 different NA subtypes. Subtypes are named by combining the H and N numbers – e.g., A(H1N1), A(H3N2). Click on the image to enlarge the picture.
There are four types of influenza viruses: A, B, C and D. Human influenza A and B viruses cause seasonal epidemics of disease (known as the flu season) almost every winter in the United States. Influenza A viruses are the only influenza viruses known to cause flu pandemics, i.e., global epidemics of flu disease. A pandemic can occur when a new and very different influenza A virus emerges that both infects people and has the ability to spread efficiently between people. Influenza type C infections generally cause mild illness and are not thought to cause human flu epidemics. Influenza D viruses primarily affect cattle and are not known to infect or cause illness in people.
Influenza A viruses are divided into subtypes based on two proteins on the surface of the virus: hemagglutinin (H) and neuraminidase (N). There are 18 different hemagglutinin subtypes and 11 different neuraminidase subtypes (H1 through H18 and N1 through N11, respectively). While there are potentially 198 different influenza A subtype combinations, only 131 subtypes have been detected in nature. Current subtypes of influenza A viruses that routinely circulate in people include: A(H1N1) and A(H3N2). Influenza A subtypes can be further broken down into different genetic “clades” and “sub-clades.” See the “Influenza Viruses” graphic below for a visual depiction of these classifications.
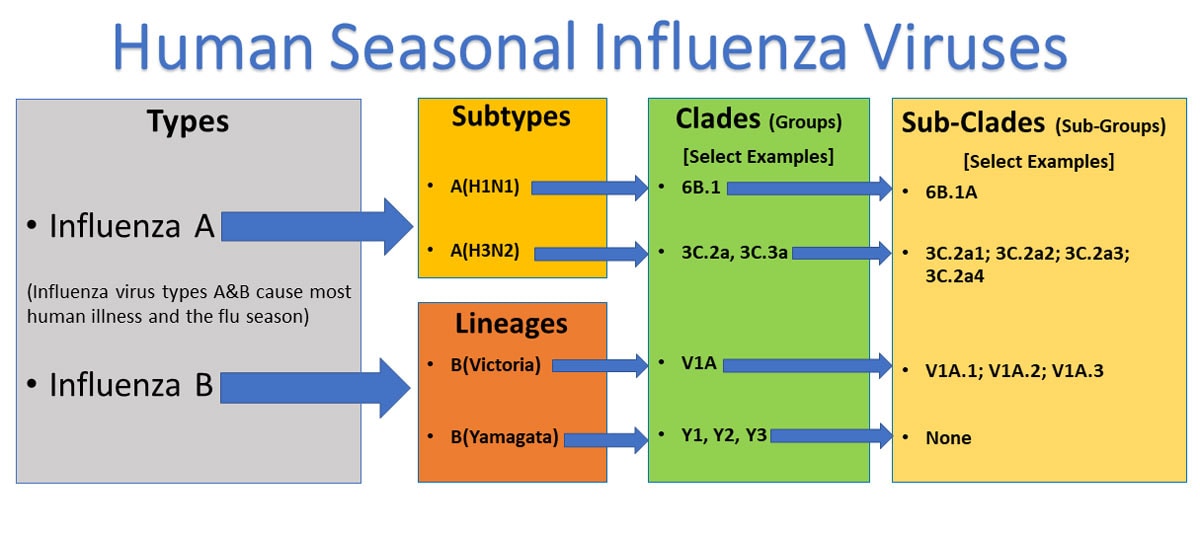
This graphic shows the two types of influenza viruses (A,B) that cause most human illness and that are responsible for the flu season each year. Influenza A viruses are further classified into subtypes, while influenza B viruses are further classified into two lineages: B/Yamagata and B/Victoria. Both influenza A and B viruses can be further classified into specific clades and sub-clades (which are sometimes called groups and sub-groups).
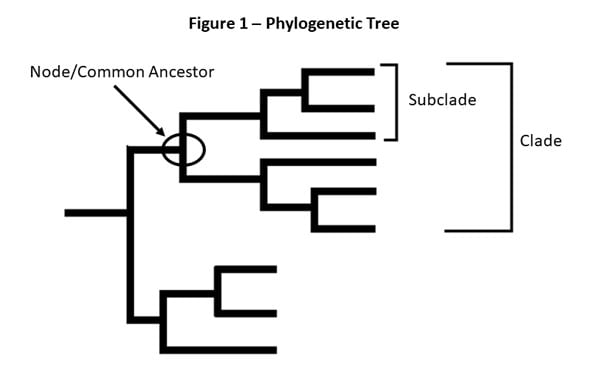
Figure 1 – This is a picture of a phylogenetic tree. In a phylogenetic tree, related viruses are grouped together on branches. Influenza viruses whose HA genes’ share the same genetic changes and who also share a common ancestor (node) are grouped into specific “clades” and “sub clades.” These clades and sub-clades are alternatively sometimes called “groups” and “sub-groups.”
Clades and sub-clades can be alternatively called “groups” and “sub-groups,” respectively. An influenza clade or group is a further subdivision of influenza viruses (beyond subtypes or lineages) based on the similarity of their HA gene sequences. (See the Genome Sequencing and Genetic Characterization page for more information). Clades and subclades are shown on phylogenetic trees as groups of viruses that usually have similar genetic changes (i.e., nucleotide or amino acid changes) and have a single common ancestor represented as a node in the tree (see Figure 1). Dividing viruses into clades and subclades allows flu experts to track the proportion of viruses from different clades in circulation.
Note that clades and sub-clades that are genetically different from others are not necessarily antigenically different (i.e., viruses from a specific clade or sub-clade may not have changes that impact host immunity in comparison to other clades or sub-clades).
Currently circulating influenza A(H1N1) viruses are related to the pandemic 2009 H1N1 virus that emerged in the spring of 2009 and caused a flu pandemic (CDC 2009 H1N1 Flu website). This virus, scientifically called the “A(H1N1)pdm09 virus,” and more generally called “2009 H1N1,” has continued to circulate seasonally since then. These H1N1 viruses have undergone relatively small genetic changes and changes to their antigenic properties (i.e., the properties of the virus that affect immunity) over time.
Of all the influenza viruses that routinely circulate and cause illness in people, influenza A(H3N2) viruses tend to change more rapidly, both genetically and antigenically. Influenza A(H3N2) viruses have formed many separate, genetically different clades in recent years that continue to co-circulate.
Influenza B viruses are not divided into subtypes, but instead are further classified into two lineages: B/Yamagata and B/Victoria. Similar to influenza A viruses, influenza B viruses can then be further classified into specific clades and sub-clades. Influenza B viruses generally change more slowly in terms of their genetic and antigenic properties than influenza A viruses, especially influenza A(H3N2) viruses. Influenza surveillance data from recent years shows co-circulation of influenza B viruses from both lineages in the United States and around the world. However, the proportion of influenza B viruses from each lineage that circulate can vary by geographic location.
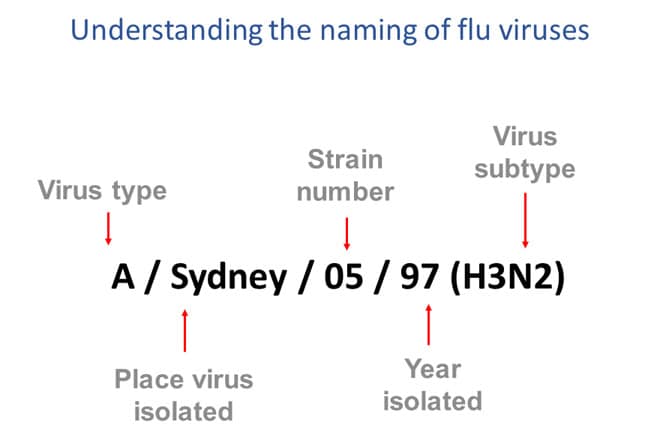
Figure 3 – This image shows how influenza viruses are named. The name starts with the virus type, followed by the place the virus was isolated, followed by the virus strain number, the year isolated, and finally, the virus subtype.
Naming Influenza Viruses
CDC follows an internationally accepted naming convention for influenza viruses. This convention was accepted by WHO in 1979 and published in February 1980 in the Bulletin of the World Health Organization, 58(4):585-591 (1980) (see A revision of the system of nomenclature for influenza viruses: a WHO Memorandum [854 KB, 7 pages]). The approach uses the following components:
- The antigenic type (e.g., A, B, C, D)
- The host of origin (e.g., swine, equine, chicken, etc.). For human-origin viruses, no host of origin designation is given. Note the following examples:
- (Duck example): avian influenza A(H1N1), A/duck/Alberta/35/76
- (Human example): seasonal influenza A(H3N2), A/Perth/16/2019
- Geographical origin (e.g., Denver, Taiwan, etc.)
- Strain number (e.g., 7, 15, etc.)
- Year of collection (e.g., 57, 2009, etc.)
- For influenza A viruses, the hemagglutinin and neuraminidase antigen description are provided in parentheses (e.g., influenza A(H1N1) virus, influenza A(H5N1) virus)
- The 2009 pandemic virus was assigned a distinct name: A(H1N1)pdm09 to distinguish it from the seasonal influenza A(H1N1) viruses that circulated prior to the pandemic.
- When humans are infected with influenza viruses that normally circulate in swine (pigs), these viruses are call variant viruses and are designated with a letter ‘v’ (e.g., an A(H3N2)v virus).
Influenza Vaccine Viruses
One influenza A(H1N1), one influenza A(H3N2), and one or two influenza B viruses (depending on the vaccine) are included in each season’s influenza vaccines. Getting a flu vaccine can protect against flu viruses that are like the viruses used to make vaccine. Information about this season’s vaccine can be found at Preventing Seasonal Flu with Vaccination. Seasonal flu vaccines do not protect against influenza C or D viruses. In addition, flu vaccines will NOT protect against infection and illness caused by other viruses that also can cause influenza-like symptoms. There are many other viruses besides influenza that can result in influenza-like illness (ILI) that spread during flu season.


































No hay comentarios:
Publicar un comentario