
The Human Protein Atlas: An Expression Map of the Complete Human Proteome
In 2003, Swedish researchers initiated the Human Protein Atlas project. This project was led by Professor Mathias Uhlén and supported by the Knut and Alice Wallenberg foundation. It is an exclusive world-leading effort, in which systematic investigation of the human proteome is carried out with the help of antibodies. An entire map of expression of proteins in all major tissues and organs in the human body is presented by the Human Protein Atlas.
The Human Protein Atlas project has produced an expression map of the entire human proteome. In order to realize this, highly specific premium antibodies are created for all protein-coding human genes, and tissue arrays are used to establish protein profiling in many cells and tissues. Applications applied are immunofluorescence staining on human cell lines (ICC-IF), Western blot analysis (WB), and immunohistochemistry (IHC). An analysis was also carried out on a subset of antibodies in HeLa cell lines, steadily expressing GFP-tagged target protein, and with the help of siRNA, a number of antibodies were further validated. Application-specific validation provides expression data.
Human Protein Atlas and Triple A Polyclonals
In order to obtain the very highest level of specificity, versatility, and reproducibility, the antibodies developed within the Human Protein Atlas project are meticulously engineered and manufactured. Early on in the project, researchers worldwide have shown a huge interest to obtain the developed antibodies. Therefore, Atlas Antibodies aims to commercialize the antibodies and make them accessible to the research community.
The Tissue Atlas and the Human Proteome
The Human Protein Atlas represents a tissue-based map of the human proteome; this was completed in 2014 following 11 years of research. The public can access all protein expression profiling data in an interactive database, allowing tissue-based analysis of the human proteome.

A tissue-based map of the complete human proteome
The Human Protein Atlas presents the tissue-based map, which is a synopsis of the present understanding of the human proteome. Furthermore, the human protein-coding genes have been divided based on their protein expression in all key tissue and organ types in the human body. In-depth information about localization and expression of all characterized human proteins is presented from seven different viewpoints, such as tissue specificity, druggability, cancerogenicity, and regulation.
The study revealed an unexpected finding that humans have relatively few tissue-specific proteins. Nearly 50% of all proteins are classified as housekeeping proteins that are expressed in all cells. It has been identified that the human body contains only 2,300 tissue-enriched proteins, that is, they have increased expression levels in specific tissues.
In order to construct the expression profiles, antibodies against human gene products are used for staining tissue micro arrays, also known as TMAs, that contain samples from 20 different cancer types, 44 different normal human tissues, and 44 different human cell lines. The expression profiles for all the proteins represented on the Tissue Atlas are based on immunohistochemistry. All protein expression data is presented in relation to RNA sequencing data for individual genes.
The 44 normal tissues exist in triplicate samples and are annotated in 76 different cell types. Moreover, all normal tissue images have been subjected to a pathology-based annotation of expression levels and are shown on the Tissue Atlas. mRNA expression data for corresponding tissue is displayed next to the protein expression data. This mRNA expression data is obtained from deep sequencing of RNA, also known as RNA-Seq, from 37 key different normal tissue types, while the protein data covers more than 17,000 genes (85%) for which antibodies are available.
To complement the immunohistochemically stained tissues, the Tissue Atlas contains the Atlas of the Mouse Brain with immunofluorescently stained tissues from different cell regions, types, and subfields of the mouse brain. The genes, which are represented on the Mouse Brain Atlas, are chosen according to function, differential- global-, and cellular expression which might not be easy to cover in the human brain.
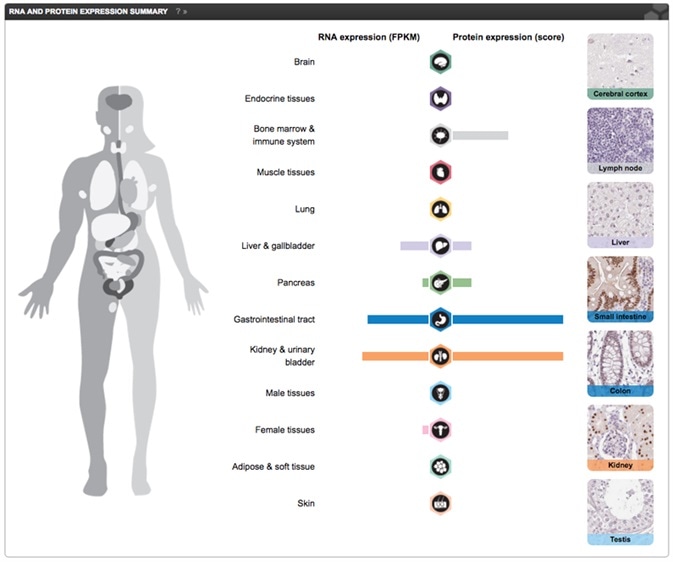
Part of the Tissue Atlas view showing RNA expression displayed together with a knowledge-based annotation, based on the IHC stainings for the particular protein, in normal tissues. The Primary data view displays the raw data from the IHC stainings in 44 normal tissues. These can be viewed in high resolution by clicking on the images. Screenshot from the Human Protein Atlas HNF1A gene Tissue Atlas entry.
The Cell Atlas and the Human Cell
The Cell Atlas, a map of the human cell, was created by the Human Protein Atlas. This atlas reveals the human cellular proteome that is localized to structures, sub-structures, and organelles in the cell.
This incredible work was accomplished through high-resolution confocal microscopy of cell lines using antibodies against more than 12,000 proteins. The Human Protein Atlas has successfully mapped the subcellular location of more than 12,000 human proteins to 32 entirely different subcellular structures. Grouping all the proteins localized to a certain structure made it possible to define the proteomes of 13 key organelles.
The study provided new interesting insights into what exactly goes on within the cell. In this context, one surprising finding was that half of the proteins were detected in multiple cellular compartments.
Subcellular localization by immunofluorescence-based confocal microscopy is presented by the Cell Atlas. Studies were conducted in three cell lines of varying origins, and the outcomes are shown as multicolor, high-resolution images of immunofluorescently stained cells. For the immunofluorescence analysis, three human cell lines for each antibody are chosen. A pair of cell lines from a human cell line panel is selected based on RNA sequencing data. U-2 OS is invariably the third cell line.
A Tool for Proteome Research
For the first time, the exclusive method adopted by the Human Protein Atlas project helps to deal with certain basic biological questions like: How large is the fraction of the human genome that encodes for cell type-specific proteins with regards to housekeeping proteins, or proteins that are differentially expressed across various cell types? How does this relate to defined cell functions, and morphological and molecular pathways? Thanks to the Human Protein Atlas, tools are now available to additionally study and interpret human biology.
Dictionaries for Learning About Histology and Cell Structure
The dictionaries are handy tools that are available at the Human Protein Atlas. Their aim is to enable the understanding and application of the information available in the Human Protein Atlas, and also serve as important tools for learning and interpreting human tissue pathology, histology, and cell biology. There are four parts in the dictionary — the Cancer Histology, Normal Tissue Histology, Protein Expression, and Cell Structure.
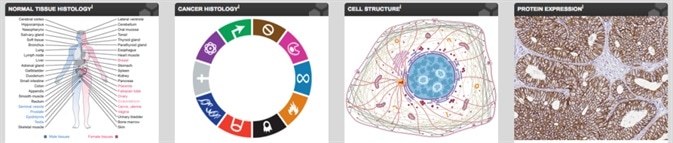
The normal histology section of the dictionary is based on representative sections of human tissues; these sections are shown in three levels of magnification. The cancer dictionary is based on representative hematoxylin-eosin (HE) stained sections from clinical tumor tissues. Organelle structure is described in the Cell structure dictionary, with immunofluorescence and high-resolution confocal microscopy images. The protein expression dictionary displays IHC-stained tissues of well-known protein and cell type markers.
Atlas Antibodies
Atlas Antibodies provides the antibodies from the Human Protein Atlas. We offer a wide range of highly validated research antibodies – Triple A Polyclonals and PrecisA Monoclonals – targeting over 75% of the human proteome.
Triple A Polyclonals are primary antibodies developed and characterized within the Human Protein Altas project. The 21,000 rabbit polyclonal antibodies cover 75% of the human proteome and are validated in IHC, WB and ICC-IF. For each IHC antibody, you can explore 500 IHC images for each antibody.
PrecisA Monoclonals are primary mouse monoclonals with unique antigen design for optimal specificity. They are epitope mapped with defined specificity and isotyped for multiplexing.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last updated: Oct 29, 2018 at 10:24 AM


































No hay comentarios:
Publicar un comentario