
Volume 25, Number 6—June 2019
Research Letter
Schistosome Interactions within the Schistosoma haematobium Group, Malawi
On This Page
Figures
Altmetric
Bonnie L. Webster, Mohammad H. Alharbi, Sekeleghe Kayuni, Peter Makaula, Fenella Halstead, Rosie Christiansen, Lazarus Juziwelo, Michelle C. Stanton, E. James LaCourse, David Rollinson, Khumbo Kalua, and J. Russell Stothard
Abstract
Molecular analysis of atypical schistosome eggs retrieved from children in Malawi revealed genetic interactions occurring between human (Schistosoma haematobium) and livestock (S. mattheei and S. bovis) schistosome species. Detection of hybrid schistosomes adds a notable new perspective to the epidemiology and control of urogenital schistosomiasis in central Africa.
Urogenital schistosomiasis is a waterborne disease transmitted by certain freshwater snails that occurs throughout much of sub-Saharan Africa. Until recently, this disease was attributed solely to Schistosoma haematobium, which was considered to have limited zoonotic potential (1). However, genetic analysis of natural infections with noninvasive larval sampling (2) has provided new evidence. In West Africa, for example, species interactions with hybrid combinations of S. haematobium and the bovine or ovine species of S. bovis and S. curassoni are commonly encountered in humans and snails (3). Although key biologic features of hybrids may not always be apparent, the risk for zoonotic transmission along with enhanced definitive and intermediate host compatibilities needs investigation (2,3). The recent emergence and persistent transmission of S. haematobium–bovis hybrids on the Mediterranean island of Corsica (4) demonstrates the public health impact of such genetic introgression.
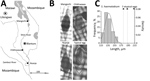
Genetic analysis of S. haematobium group species in central and southern Africa is a high priority. Atypical egg morphologies suggest a capacity for natural hybridization of S. haematobium with the bovine species S. mattheei, later confirmed with biochemical markers and experimental infections demonstrating viable progeny (3). During ongoing surveillance of urogenital schistosomiasis in Chikhwawa District, Malawi, we encountered atypical S. haematobium eggs in urine samples from several infected children (5). We report the further genetic characterization of atypical eggs collected from epidemiologic surveys of children within Chikhwawa, Nsanje, and Mangochi Districts (Figure, panel A).
Ethics approvals for the epidemiological surveys were granted by Liverpool School of Tropical Medicine; College of Medicine, Malawi; and Ministry of Health and Population, Malawi. All children found infected were treated with praziquantel.
We filtered schistosome eggs from the urine of infected children, then photographed and measured them before storing them on Whatman FTA cards for molecular analysis (2). We alkaline-eluted and genotyped DNA from individual eggs using both the mitochondrial cytochrome oxidase subunit 1 (cox1) and the nuclear ribosomal internal transcribed spacer (rITS) DNA regions (2) (Appendix Table). In addition, for the samples from Mangochi District, we analyzed a partial region (300-bp) of the nuclear ribosomal 18S DNA to confirm the presence of S. mattheei nuclear DNA (2,6) (Appendix).
Of 6 atypical eggs from Chikhwawa, all had a pure S. haematobium genetic profile (Figure, panels B, C). Of 19 eggs from Nsanje, 18 had a pure S. haematobium genetic profile; 4 eggs had atypical morphology, but only 1 atypical egg had a discordant genetic profile (i.e., cox1 S. bovis and rITS S. haematobium). Of 20 eggs from Mangochi, 16 typical S. haematobium eggs had a pure S. haematobium genetic profile, whereas the 4 atypical eggs had the same discordant genetic profiles (cox1 S. mattheei and rITS S. haematobium-mattheei). Inspection of the partial 18S gene sequence confirmed S. haematobium–mattheei hybrids (Appendix). We deposited all sequence data into GenBank (accession nos. MK358841–MK358858).
Our genetic analysis demonstrated the presence of S. haematobium group hybrids in Malawi as introgressed forms of S. haematobium–mattheei and S. haematobium–bovis. Of note, an unusual egg morphology may not always correspond with the ability to detect introgression with the current combination of genetic markers used (6; Appendix). As described by Boon et al., successive backcrossings of hybrid progeny may obscure our ability to detect ancestral introgression, and the development of a wider panel of nuclear genetic markers is needed (6). Nonetheless, detection of these 2 hybrid schistosomes strongly suggests interactions of S. haematobium with the ungulate schistosomes S. mattheei and S. bovis. That S. bovis has not been reported in Malawi implies a changing species dynamic with possible zoonotic transmission along the drainage basin of Lake Malawi, adding a new dimension to the epidemiology and control of urogenital schistosomiasis in Malawi (7).
Because we did not attempt miracidial hatching during this study, we cannot confirm that these hybrids or introgressed forms are fully viable in autochthonous natural transmission. However, the process of ancestral introgression with subsequent natural selection may help explain unexpected shifts in local snail–schistosome relationships (e.g., the changing compatibility of Bulinus nyassanus snails in Lake Malawi with S. haematobium schistosomes) (8). Further studies are needed to better characterize schistosomes involved in human infection, investigate more thoroughly any zoonotic potential, and assess all possible combinations of interspecies introgressions.
Molecular evidence for ancestral hybridization between S. haematobium and S. mansoni schistosomes was presented recently (9); given autochthonous transmission of intestinal schistosomiasis in Lake Malawi (10), there may be sufficient epidemiologic opportunity for other introgression events to occur with the hybrids we report. We therefore advise heightened concurrent surveillance of urogenital and intestinal schistosomiasis, entailing a OneHealth approach with molecular vigilance for interspecies interactions along with phenotypic assessments for any altered host pathogenicity or susceptibility to praziquantel treatment. Detection of the hybrid schistosomes we report adds a new perspective to the epidemiology and control of urogenital schistosomiasis in central Africa.
Dr. Webster is a researcher with the Natural History Museum, London, UK. She has specific expertise in medical helminthology and a longstanding interest in studies of schistosome hybridization in both natural and experimental settings.
Acknowledgments
We are particularly grateful to the local health and education authorities of Malawi, district teachers, and community health workers involved in schistosome surveys in Chikhwawa, Nsanje, and Mangochi. We thank Jahashi Nzalawahe for assistance with measurements and analysis of schistosome egg morphology.
M.H.A. is funded by a PhD scholarship from the Ministry of Health, Kingdom of Saudi Arabia. S.K. received a scholarship from Commonwealth Scholarship Commission and a research fellowship from the African Research Network for Neglected Tropical Diseases (ARNTD).
References
Figure
Cite This ArticleOriginal Publication Date: 5/3/2019





















.png)












No hay comentarios:
Publicar un comentario