
Purine Biosynthesis
Contextualizing purines
Purines are heterocyclic bases. Simply put, these are closed ring structures comprised of at least two different kinds of atoms. Purines are one of three components of nucleotides; phosphate esters of a pentose sugar (either ribose or deoxyribose) in which a purine or pyrimidine base is linked to C1 of the sugar.
The prefix mono- di- or tri- denote the number of phosphate groups present on the nucleotide. It is important to distinguish the nucleoside; this is the non-phosphorylated form of a nucleotide. It is
Nucleoside triphosphates are the monomeric units that act as precursors of nucleic acids. These perform a wide range of biochemical functions that include
- Driving thermodynamically unfavourable reactions
- Forming the central cofactors of metabolism (such as NAD+ and FAD+)
- Forming the building blocks of our genetic blueprint, DNA.
Figure 1. The structure of nucleotides depicting how the base and pentose sugar (nucleoside, in yellow blue and green) may be attached to either one, two or three phosphate groups. A nucleoside attached to one phosphate (red) is a nucleoside monophosphate.
The addition of e second phosphate group (red) forms a nucleoside diphosphate, and finally, the addition of a third phosphate forms a nucleoside triphosphate. Where the phosphate group proximal to the nucleoside the site of the phosphate-ester bond.
Structure of purines
Purine biosynthesis is complex. The purine skeleton is a 6-membered pyrimidine ring fused to a 5-membered imidazole ring (See Figure 1). Each ring contains two nitrogen (N) atoms, with the remaining 5 positions in each ring occupied by carbon (C), which is attached to a Hydrogen (H).
Hydrogen can be replaced by different atoms or groups to form distinct purines. The 4 Ns originate from different amino acids and the remaining 5 Cs derive from one-carbon containing groups.
This was discovered in 1948 by John Buchanan who fed pigeons isotopically labelled compounds to determine the positions of the labelled atoms in the uric acid they secreted. The name of the compound that provides each of the C and N atoms are labelled in Figure 1.
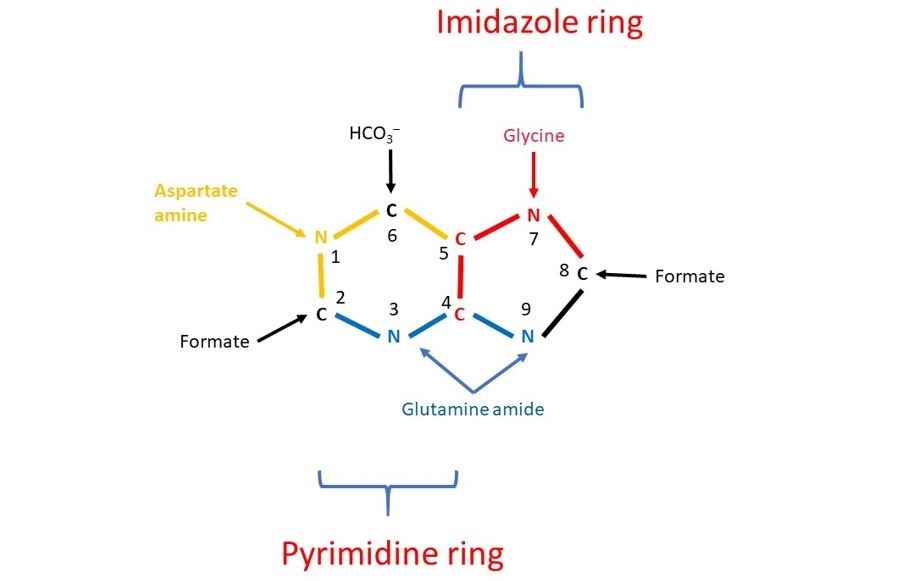
Figure 2 The results of John Buchanan’s studies demonstrated that N1 of purines arises from the amino group of aspartate; C2 and C8 originate from a C1 containing compound called formate; N3 and N9 are contributed by the amide (NH2) group of glutamine; C4, C5, and N7 descend from Glycine and C6 comes from HCO3–.
Synthesis of purine ribonucleotides
Purine biosynthesis occurs in the cytosol of all cells. The purine ring is built up in a series of 11 enzyme catalysed steps. Each enzyme is oligomeric, which means it contains several monomers. Intermediate products that are produced during the reaction are not released. Instead, they are shuttled to the subsequent enzyme along the pathway.
Step one of this pathway generates an important compound, 5-phosphoribosyl-alpha-pyrophosphate (PRPP). This compound is also a precursor in the biosynthesis of pyrimidine nucleotides. It provides the phospho-ribose units of these ribonucleotides.
PRPP is derived from ribose-5-phosphate (R5P), a product of the pentose phosphate pathway. Therefore, purines are built from a series of addition reactions to a sugar.
Purine synthesis yields inosine monophosphate
In the first step of purine biosynthesis, ribose phosphate pyrophosphokinase activates the ribose by reacting it with ATP to form 5-phosphoribosyl-alpha-pyrophosphate (PRPP).
Step 2 is the committed step of purine biosynthesis. In this reaction amidophosphoribosyl transferase catalyses the displacement of PRPP’s pyrophosphate group by glutamine’s amide nitrogen. This reaction is the pathway’s flux-controlling step i.e. the rate at which the biosynthetic pathway outputs product. It is shown in figure 3.
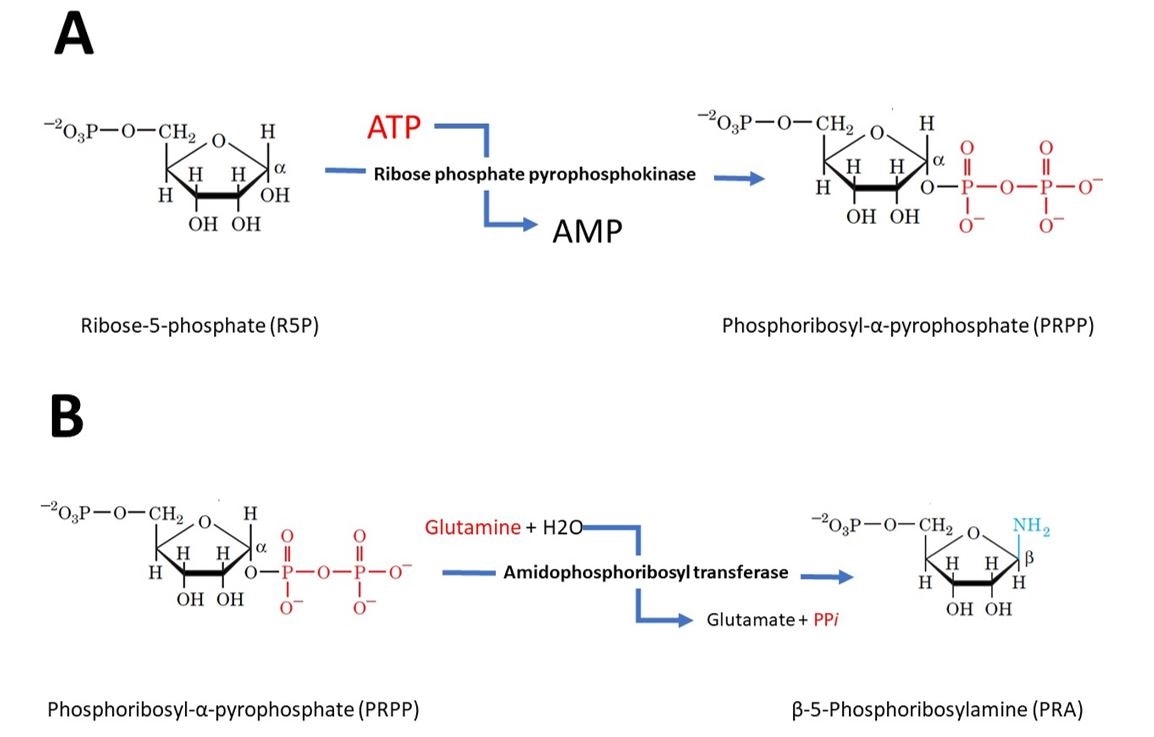
Figure 3 (A) Step 1 - Activation of ribose-5-phosphate. The starting material for purine biosynthesis ribose-5-phosphate, a product of the pentose phosphate Pathway. In the first step of purine biosynthesis, ribose phosphate pyrophosphokinase activates the ribose by reacting it with ATP, which drives the reaction, to form 5-phosphoribosyl-alpha-pyrophosphate (PRPP). (B) Step 2 – Flux-controlling step. Amidophosphoribosyl transferase catalyses the displacement of PRPP’s pyrophosphate group by glutamine’s amide nitrogen forming Beta-5-phosphoribosylamine. This step is also driven by ATP.
Following the remaining 9 steps, the first purine derivative that is synthesised is inosine monophosphate (IMP). This can be seen in Figure 4.
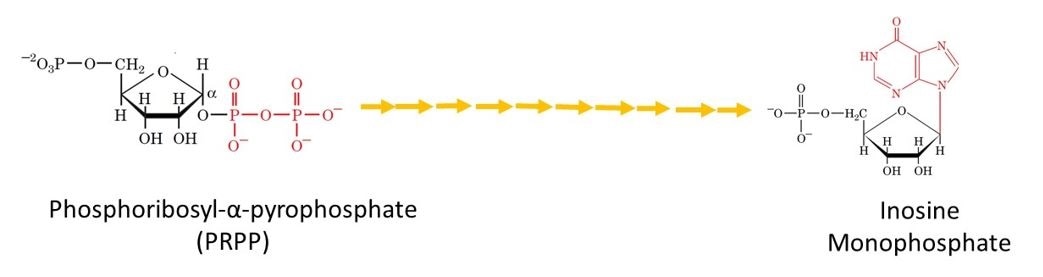 Figure 4. The metabolic pathway for the de novo biosynthesis of IMP. Here the purine residue is built up on a ribose ring in 11 enzyme catalyzed reactions.
Figure 4. The metabolic pathway for the de novo biosynthesis of IMP. Here the purine residue is built up on a ribose ring in 11 enzyme catalyzed reactions.IMP is the precursor for the purine nucleotide, adenosine and guanosine monophosphate (AMP and GMP). Each are synthesized in a two-reaction pathway with bifurcates at the level of IMP:
Further phosphate additions to generate diphosphate and triphosphate nucleosides may follow completion of monophosphate synthesis. These reactions are carried out by kinases.
Kinases are so-called due to their property of transferring phosphate groups from a high-energy phosphate molecule to specific substrates. The complete nucleotide triphosphate forms, adenosine and guanosine triphosphate (ATP and GTP) are the recognizable units of RNA and DNA. Therefore, purines are initially formed as ribonucleotides rather than as free bases.
Purine nucleotide biosynthesis is regulated at several steps
The pathways synthesizing IMP, ATP, and GTP are individually regulated. This is crucial to prevent the waste of (1) energy and nitrogen, (2) to control the total amounts of purine nucleotides available for nucleic acid synthesis and (3) the purine waste product, uric acid, is are harmful to cells. Excessive uric acid production leads to its deposition in joints causing pain and redness; this the pathophysiological basis of gout.
IMP synthesis is controlled by the levels of adenine and guanine nucleotides. Additional control is exerted by feedforward activation, which is the stimulation of a subsequent enzyme by the preceding substrate. In this situation, the amidophosphoribosyl transferase I step 2 is allosterically stimulated by PRPP, the product of step 1.
The second level of regulation occurs at the branch point below IMP, leading to either AMP or GMP. These end products are each competitive inhibitors of IMP and so, their excessive build-up is prevented.
The metabolic requirement for purine can be fulfilled by biosynthesis in the human body. Without adequate production of purines, or because of abnormal biosynthetic pathways, painful clinical manifestations can arise.
Sources
Further Reading
Last Updated: Oct 16, 2018


































No hay comentarios:
Publicar un comentario