
New protein sequencing technique could revolutionize biomedical research
Researchers at The University of Texas at Austin have demonstrated a new approach to protein sequencing that is much more sensitive than the current technique.
The new technique allows individual proteins to be identified, rather than requiring millions of them to be determined simultaneously.
The development could have major implications for biomedical research, making it easier to detect new biomarkers for disease, as well as improving understanding of how healthy tissue functions.
The advancement is the result of work started six years ago by Dr. Edward Marcotte and colleagues, who were interested in adapting the methods used in next generation sequencing (NGS) to protein sequencing.
In the same way that NGS enabled fast and accurate sequencing of the entire genome, the technology developed by the team provides quick and detailed information about tens of thousands of proteins involved in health and disease.
In many diseases such as cancer, diabetes and Alzheimer’s, cells produce proteins that serve as unique fingerprints or biomarkers for the disease.
Improving detection of these biomarkers would help researchers better understand the diseases and enable earlier and more accurate diagnosis.
The current standard for protein sequencing – mass spectrometry – is often not sensitive and requires around one million copies of a protein to be present in order for it to be detected.
The technique is also “low throughput,” detecting only a few thousand types of protein in a single sample.
As reported in the journal Nature Biotechnology, the new method, called single molecule fluorosequencing, enables millions of individual proteins to be sequenced simultaneously in a single sample.
With further refinement of the technique, Marcotte is hopeful that this number could be increased to reach into the billions.
The higher throughput and sensitivity of the new technique compared with the current standard should improve the detection of disease biomarkers and enable scientists to study diseases such as cancer in an entirely new way.
Looking at a tumor an a single-cell level, for example, could enable researchers to see how a tumor evolves form a small collection of identical cells to a large mass of genetically diverse cells with different properties. Such insights could pave the way for new approaches to targeting cancer.
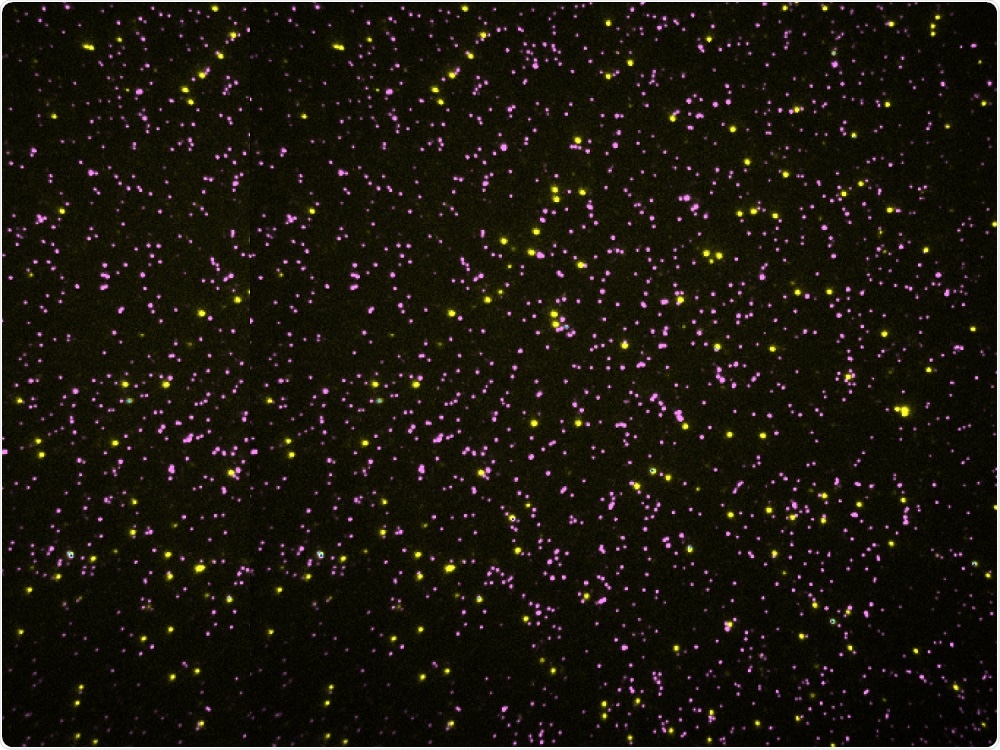 Credit: University of Texas at Austin
Credit: University of Texas at AustinThe dots in this picture aren't stars, they're millions of proteins as seen through a microscope. Each protein stands up like a blade of grass with only the tips visible as we look down from above.
Proteins are chains of amino acids, so each dot is just the amino acid on the tip. Of the 20 flavors of amino acids that make up proteins, one type is tagged yellow, another is tagged pink, the other 18 aren't tagged, they're black.
By removing one amino acid from each protein, taking a new picture and then repeating many times, researchers can record a sequence of amino acids for each protein in a sample of millions simultaneously.
Source:
This news story is based on a press release by The University of Texas at Austin.



































No hay comentarios:
Publicar un comentario