
Gut-on-a-chip to model coeliac disease
 By Keynote ContributorProf. Cisca WijmengaLodewijk Sandkuijl Endowed Chair,
By Keynote ContributorProf. Cisca WijmengaLodewijk Sandkuijl Endowed Chair,Professor of Human Genetics,
University Medical Centre Gronigen (UMCG)
Coeliac disease is the most common food intolerance in the Western world affecting 1% of the population. Coeliac disease is characterized by a chronic immune reaction in the small intestine resulting from an aberrant cellular response to our diet. The classical presentation of the disease is in infants with diarrhea and failure to thrive. However, nowadays the disease is often diagnosed in adults and often remains undiagnosed probably because the disease can present in many different ways, also outside the gastrointestinal tract.
nobeastsofierce | Shutterstock
There are many serious associated health complications if coeliac disease is left undiagnosed, including growth problems and delayed puberty in children, iron deficiency anaemia, chronic fatigue and osteoporosis. The lack of awareness of coeliac disease, in both members of the public and healthcare professionals, means that the diagnosed cases of the disease only represent a small fraction of the total number of people affected.
Common symptoms of coeliac disease include abdominal bloating and pain, diarrhoea, constipation, vomiting, mouth ulcers, fatigue and anaemia. A significant challenge in recognising coeliac disease is the variation in the presentation and intensity of symptoms and, in many cases, coeliac disease may even occur without any symptoms.
Gluten is a storage protein present in wheat, barley and rye, and is needed to maintain the processing quality of food derivates such as bread, pasta, and cookies. Coeliac disease starts in the small intestine where breakdown products of dietary gluten, like many other dietary products, pass the epithelial lining.
However, instead of being recognized as a normal food product these breakdown products are seen as “foreign” and elicit an unwanted immune response. In the intestine, gluten is partially degraded and modified, which results in gluten peptides that bind with to HLA-DQ2 or HLA-DQ8 molecules and trigger an adaptive inflammatory T cell response.
At the same time, gluten exposure leads to an innate immune response characterized by increased production of IL15 by intestinal epithelial cells, leading to epithelial cell destruction. The link with dietary gluten present in wheat, rye and barley has been established in the early 1950s of the previous century. Hence, the only treatment today is a lifelong gluten-free diet.
Coeliac disease is a clear genetic disorder as first-degree relatives of patients are also at increased risk for disease development. However, the disease is multifactorial and involves multiple genetic risk factors together with non-genetic risk factors. Over the past 40 years it has become clear that a particular genetic make-up of the major histocompatibility complex (MHC) – cell surface molecules recognizing foreign molecules – is a prerequisite to develop coeliac disease.
Possession of these so-called HLA-DQ2 molecules are also common in the healthy population, suggesting that there are other genetic risk factors that also contribute to disease etiology. Over the past 10 years we have identified at least 40 more genes in the human genome that also increase risk to develop celiac disease.
However, the interaction between HLA and gluten peptides take place after the gluten breakdown products have passed the epithelial lining and it is still unclear how this passage takes place and whether there is already an effect of gluten breakdown products at the epithelial surface itself.
Answering these questions requires a model system that recapitulates the small intestine of coeliac disease patients including the correct genetic background that predisposes to coeliac disease. Many of the genes that increase risk for coeliac disease influence the immune system but some genes may affect the barrier function of the intestinal epithelial lining.
It might be hypothesized that these barrier genes somehow affect the transport of gluten breakdown products. Such a complex genetic background cannot be modeled in animal models like mice. Therefore, we a need a system where the starting point includes cells from coeliac disease patients. This motivated us to develop a gut-on-a-chip which would overcome the limitations of animal and cell-based models.
A gut-on-a-chip mimics the properties of the human gut and in our case those of coeliac disease patients. Using blood or urine we are able to derive induced pluripotent stem cells which in turn can be differentiated into intestinal epithelial cells.
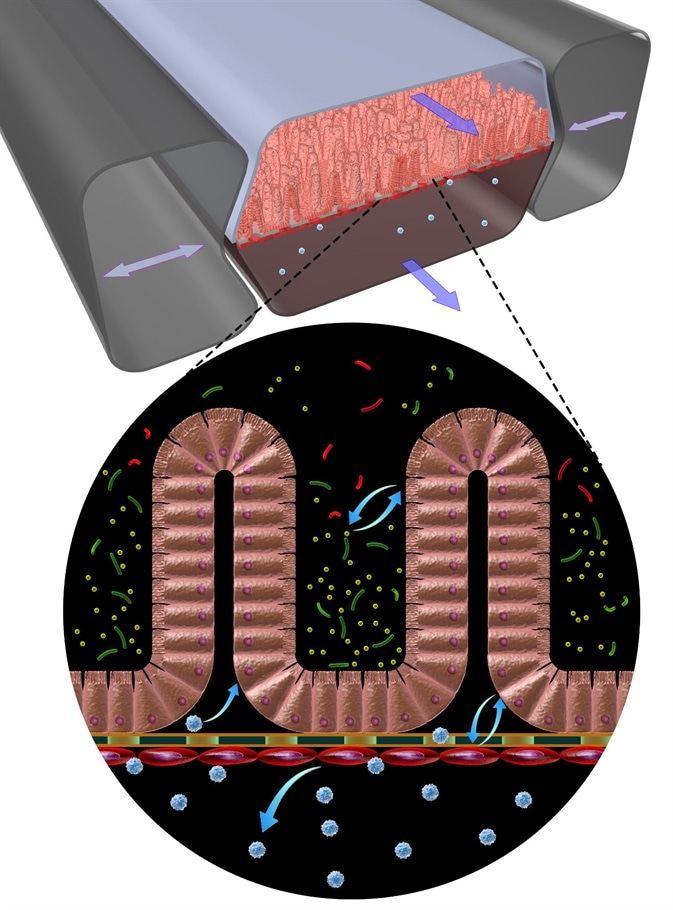
Credit Wyss Institute at Harvard University
These living, in vitro models adapt a villus architecture very similar to what is normally seen in the small intestine and that can absorb and transport molecules. With such a system in hand we can study for example the transport of gluten breakdown products in the presence or absence of a particular genetic backgrounds.
Using Crisp/Cas9 technology we can also perform gene-editing experiments and completely remove genes to test their involvement in the disease process. In addition, we can study if other factors like infections or the gut microbiome, also affect the barrier function of the epithelial layer and have an effect on the immune response in coeliac disease.
The gut-on-a-chip will be crucial to increase our basic understanding of the disease etiology. At the same time, it may also help to find diagnostic marker which can help to identify patients-at-risk easier and more frequently. Or to find prognostic markers that can be used to monitor adherence to a gluten-free diet, something that is extremely difficult at the moment.
Of the genetic risk factors that we identified for coeliac disease about 30% are potential drug targets. Having a gut-on-a-chip we may also use such as systems to test if certain drugs can be used to treat coeliac disease. Finally, we may also enhance our understanding in the shared etiology between coeliac disease and other autoimmune disorders.
About Prof Cisca Wijmena
Cisca Wijmenga currently holds the position of Lodewijk Sandkuijl Endowed Chair and Professor of Human Genetics at the University Medical Centre Groningen (UMCG), the Netherlands. She is also a member of the Royal Holland Society of Sciences and Humanities (KHMW) and the Netherlands Royal Academy of Arts and Sciences (KNAW) and has recently been appointed Knight of the Order of the Netherlands Lion.
This year, Professor Cisca Wijmenga has been awarded the distinguished UEG Research Prize 2018 for her work on coeliac disease. Her research focuses on the understanding of genetic variation and how this leads to phenotypic variation, with a particular focus on its relationship to autoimmune diseases.
Disclaimer: This article has not been subjected to peer review and is presented as the personal views of a qualified expert in the subject in accordance with the general terms and condition of use of the News-Medical.Net website.
Last Updated: Oct 22, 2018



































No hay comentarios:
Publicar un comentario