
Volume 23, Number 12—December 2017
Research
Outbreaks of Neuroinvasive Astrovirus Associated with Encephalomyelitis, Weakness, and Paralysis among Weaned Pigs, Hungary
On This Page
Ákos Boros, Mihály Albert, Péter Pankovics, Hunor Bíró, Patricia A. Pesavento, Tung Gia Phan, Eric Delwart, and Gábor Reuter
Abstract
A large, highly prolific swine farm in Hungary had a 2-year history of neurologic disease among newly weaned (25- to 35-day-old) pigs, with clinical signs of posterior paraplegia and a high mortality rate. Affected pigs that were necropsied had encephalomyelitis and neural necrosis. Porcine astrovirus type 3 was identified by reverse transcription PCR and in situ hybridization in brain and spinal cord samples in 6 animals from this farm. Among tissues tested by quantitative RT-PCR, the highest viral loads were detected in brain stem and spinal cord. Similar porcine astrovirus type 3 was also detected in archived brain and spinal cord samples from another 2 geographically distant farms. Viral RNA was predominantly restricted to neurons, particularly in the brain stem, cerebellum (Purkinje cells), and cervical spinal cord. Astrovirus was generally undetectable in feces but present in respiratory samples, indicating a possible respiratory infection. Astrovirus could cause common, neuroinvasive epidemic disease.
Astroviruses are small, nonenveloped viruses with single-stranded 6.2–7.8 kb RNA genome of positive polarity (1,2). The family Astroviridae is currently divided into 2 genera: the genus Mamastrovirus of mammal-infecting viruses and the genus Avastrovirus of avian viruses (3,4). The genetically heterogenic astroviruses that are widespread among mammals and birds are generally associated with gastroenteritis, less commonly with respiratory disease, and rarely encephalitis or disseminated infections (2,5–19). Astrovirus infections with central nervous system (CNS) involvement were reported recently in mink, human, bovine, ovine, and swine hosts (the latter in certain cases of AII type congenital tremors) (5,6,12–14). Most neuroinvasive astroviruses belong to the Virginia/Human-Mink-Ovine (VA/HMO) phylogenetic clade and cluster with enteric astroviruses identified from asymptomatic or diarrheic humans and animals (15,16). Recent research shows that pigs harbor one of the highest astrovirus diversities among mammals examined (3,15,20). Porcine astroviruses (PoAstVs) were identified mainly from diarrheic fecal specimens, less commonly from respiratory specimens, although the etiologic role of astrovirus infection in gastroenteritis or in other diseases among swine is not settled (3,9,20–23). We report the detection of neuroinvasive porcine astrovirus type 3 (Ni-PoAstV-3) by reverse transcription PCR (RT-PCR) and in situ hybridization (ISH) in recent and archived CNS samples of newly weaned paraplegic pigs from 3 highly prolific swine farms in Hungary.
Sample Collection and Handling
During November 2015–July 2017, we collected multiple tissue samples from 5 paraplegic and 5 asymptomatic pigs at the index farm located in Hungary (GD; specific location redacted) (Table 1). We also tested nasal and anal swab pairs collected by using polyester-tipped swabs from another 5 paraplegic and 13 healthy animals. We washed tissue samples twice in 10 mmol/L phosphate buffered saline (PBS) to remove excess blood and held them at −80°C until total RNA extraction. For formalin-fixed, paraffin-embedded (FFPE) blocks, we fixed the dissected samples (Table 2) with buffered 8% formaldehyde, dehydrated and embedded into paraffin.
We also analyzed archived FFPE specimens from paraplegic pigs from earlier outbreaks of posterior paraplegia in Tázlár in 2011 and in Balmazújváros in 2014 (Table 2). The 3 swine farms are located in the central and eastern parts of Hungary, ≈100 km from each other, without known connection.
Previous Laboratory Diagnostics
CNS homogenates from the index farm tested negative by PCR for the following pathogens (families in parentheses): porcine reproductive and respiratory syndrome virus (Arteriviridae); porcine circovirus 2 (Circoviridae); hemagglutinating encephalitis virus (Coronaviridae); and porcine parvovirus 1, 2, 4, and porcine bocavirus (Parvoviridae). Immunohistochemical detection of Toxoplasma gondii and West Nile virus and bacterial cultivation attempts from the CNS samples were also negative. Virus isolation attempts using brain homogenates of affected animals in swine kidney (PK-15) and Caucasian colon adenocarcinoma (Caco-2) cell lines were not successful (no cytopathic effects were visible). We detected no PoAsV type 3 (PoAstV-3) in the cell culture supernatants by nested RT-PCR with RNA-dependent RNA polymerase (RdRp) primer pairs.
Total RNA Extraction and RT-PCR Screening

Figure 1. Genome map of the neuroinvasive PoAstV-3 strain NI-Brain/9-2016a/HUN (GenBank accession no. KY073229) from a symptomatic newly weaned pig from a farm in Hungary together with the location of RT-PCR products used...
Treatment of FFPE samples included the deparaffination and rehydration steps, proteinase K digestion, and total RNA extraction. We used the same treatment protocols and the same reaction conditions and reagents used in the RT-PCR and nested RT-PCR reactions as are described previously, with minor modifications (24–26) (Technical Appendix[PDF - 372 KB - 5 pages]). For the RT-PCR screening of CNS samples for the presence of pestiviruses (Flaviviridae) and swine picornaviruses (Picornaviridae), including teschovirus, enterovirus, sapelovirus, Seneca Valley virus, pasivirus, kobuvirus and encephalomyocarditis, we used virus-specific primer pairs as well as the outer and inner primer pairs targeting the RdRp or the capsid regions of PoAstV-3 (Figure 1; Technical Appendix[PDF - 372 KB - 5 pages]Tables 1, 2).
Absolute Quantification Using Quantitative RT-PCR
For the absolute quantification of viral RNA present in different tissue, urine, and fecal samples, we used the SYBR Green–based quantitative RT-PCR (RT-qPCR) method (Maxima SYBR Green qPCR Master Mix; Thermo Scientific, Waltham, MA, USA). For the generation of standard curve, we used 10-fold dilution series of purified and spectrophotometrically quantified RNA transcripts in the reactions. The RT-qPCR assays contained 3 technical repeats of all samples and standards. The slope of the standard curve was −3.4228 and the calculated PCR efficiency was 99.96%. The detailed protocol is provided in the online Technical Appendix.
Long-range Amplification, 5′/3′ RACE-PCR, and Sanger Sequencing
For the complete genome (or complete 3′ open reading frame [ORF] 1b–ORF2–3′ untranslated region [UTR]) acquisitions of the PoAstVs, we used different long-range and 5′/3′ rapid amplification of cDNA ends RT-PCRs according to previously described protocols (26,27). We designed the sequence-specific primers used for the amplification of overlapping genome fragments based on the genome of PoAstV-3 strain US-MO123 (GenBank accession no. JX556691) and closely related sequences downloaded from the GenBank database (Technical Appendix[PDF - 372 KB - 5 pages] Table 3). We sequenced PCR products directly with the BigDye Terminator v1.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Stafford, TX, USA) using the primer-walking method with an automated sequencer (ABI Prism 310 Genetic Analyzer; Applied Biosystems). We have submitted the nucleotide sequences of study astrovirus strains to GenBank under accession nos. KY073229–32.
Sequence and Phylogenetic Analyses
We aligned astrovirus sequences by using the MUSCLE web tool of EMBL-EBI (28) and performed pairwise nucleotide and amino acid identity calculations of the aligned sequences with GeneDoc version 2.7 (http://iubio.bio.indiana.edu/soft/molbio/ibmpc/genedoc-readme.html). We constructed phylogenetic trees of deduced amino acid sequence alignments by using MEGA version 6.06 software (29) and the neighbor-joining method with the Jones–Taylor–Thornton matrix-based model. Bootstrap values were set to 1,000 replicates, and only likelihood percentages of >50% were indicated.
Histology and In Situ Hybridization
We performed chromogenic (with 3,3′-diaminobenzidine/DAB) in situ hybridization in FFPE slides (RNAscope 2.0, Brown Kit; Advanced Cell Diagnostics, Newark, CA, USA) according to the manufacturer’s instructions for viral RNA detection of Ni-PoAstV-3. We used 30 probe pairs generated at Advanced Cell Diagnostics designed to hybridize native viral Ni-PoAstV-3 RNA. Negative controls included Dap-B (dihydrodipicolinate reductase gene from Escherichia coli probe); an unrelated viral probe; and normal porcine brain region-matched sections.
Clinical Observations

Video. Stage 1: Weaned pig with posterior weakness. In milder cases and in early stage of the disease, lesions are confined to the lumbar part of the spinal cord, resulting in posterior...
There are ≈2,000 sows and their offspring in the investigated highly prolific index farm (GD). Episodes of neurologic disease of unknown etiology have persisted in the past 2 years. The syndrome affects an average of 30–40 weaned pigs monthly (1.5%–2% of total), although the number of monthly cases infrequently rose to ≈80 pigs (4%) in the autumn–winter seasons. The clinical signs of posterior leg weakness or paraplegia and pitching (stage 1); later paralysis of both legs and skin pain (stage 2); or loss of consciousness, paresis, and serious flaccid paralysis of muscles (stage 3) typically appear among weaned pigs 25–35 days old, 1 week after the weaning procedure (Video). We did not observe gastroenteric symptoms. All of the affected pigs in stage 3 of the disease were unable to eat or drink; they died due to exsiccosis (dehydration) or were euthanized. Signs persisted typically for 1 week before death or euthanasia. Postmortem examination results showed no signs of mechanical damage (fractures, abscesses, or hemivertebrae). Pigs are vaccinated against porcine circovirus 2, Mycoplasma hyopneumoniae, and Actinobacillus pleuropneumoniae. Preventive amoxicillin treatment of the piglets was done routinely at weaning. Due to the preventive measures in effect as of spring 2017, which included extensive decontamination of the piggeries and the physical separation of the newly weaned pigs from different litters, the number of encephalomyelitis cases among weaned pigs decreased with only 1–2 cases/month observed on the index farm.
The 2 additionally examined swine farms located in Tázlár and Balmazújváros each held approximately 500 sows and their offspring. Similar symptoms of staggering and paralysis appeared among pigs 3–5 weeks old in outbreaks in 2011 (Tázlár) and 2014 (Balmazújváros).
Detection and Analysis of Astroviruses from CNS Samples of Affected Animals
In March 2016, we collected brain stem, spinal cord, nasal swab, and fecal samples from a newly weaned pig from index farm GD (GD-1, index animal) that showed signs of encephalomyelitis and posterior paraplegia (stage 1). The brain stem and spinal cord samples tested negative by RT-PCR for pestivirus (family Flaviviridae) and several swine-infecting picornaviruses (family Picornaviridae) (Technical Appendix[PDF - 372 KB - 5 pages] Table 1). On the basis of the increasing evidence of the pathogenic role of neurotropic astroviruses among humans and farm animals (5,6,14,17,30) we investigated the presence of astrovirus using panastrovirus PCR primers (Technical Appendix[PDF - 372 KB - 5 pages] Table 1) (31). The brain stem and spinal cord samples showed strong RT-PCR positivity. The panastrovirus PCR products were sequenced using panastrovirus PCR primers (Technical Appendix[PDF - 372 KB - 5 pages] Table 1) and compared to each other and to the available astroviruses using blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The 397-nt sequences of brain stem and spinal cord were identical and showed 89% nt identity to PoAstV-3 isolate US-MO123 (GenBank accession no. JX556691) as the closest match (32).
We sequenced 2 samples from the index animal: the full-length genome of the neuroinvasive astrovirus strain NI-Brain/9-2016a/HUN (GenBank accession no. KY073229) from the brain stem sample and the complete capsid-encoding ORF2 from the spinal cord sample NI-SC/9–2016a/HUN (GenBank accession no. KY073230). The 6393-nt (without the poly[A] tail) complete genome showed the typical astrovirus genome organization with 3 putative ORFs, 2529 nt (ORF1a), 1527 nt (ORF1b), and 2265 nt (ORF2), flanked by short 5′ and 3′ UTRs (Figure 1). We identified the conserved proteolytic cleavage site (V561HQ↓TNT) of serine protease (ORF1a) and the conserved Y358GDD motif of the RdRp (ORF1b) (33). The nonstructural proteins of ORF1a (842 aa) and ORF1b (508 aa) and the capsid protein of ORF2 (754 aa) showed 93%, 95%, and 93% aa identity, respectively, to the corresponding genome parts of the closest known relative PoAstV-3 strain, US-MO123. All of the conserved genomic features of mamastroviruses were present in strain NI-Brain/9-2016a/HUN: the conserved C1CAAA pentamer at the 5′ end of the genome; the frame-shift heptamer motif (A2511AAAAAC) followed by a stem–loop structure at the 3′ end of ORF1a; the conserved sgRNA promoter sequence motif of U4048UUGGAGgGGaGGACCaAAN8AUGgC (variable nts are in lowercase, start codon of ORF2 is underlined) at the junction of ORF1b/ORF2; and the stem loop II-like motif (s2m) in the 3′ end of the genome between nt positions 6322 and 6353. The 3′ UTR of NI-Brain/9-2016a/HUN is 27 nt shorter and did not contain the short sequence repeat found at the 3′ end of strain US-MO123 (G6381/6392AUUUCUUUNA). Based on the high sequence identity and the similar genomic features, the NI-Brain/9-2016a/HUN strain most likely belongs to the PoAstV-3 genotype. The ORF2 of NI-Brain/9-2016a/HUN shares 99% nt/aa identity with the corresponding capsid gene of NI-SC/9-2016a/HUN from the spinal cord of the same animal, suggesting that the same virus was present in both regions of the CNS.
We detected Ni-PoAstV-3 using RT-PCR in all CNS samples collected from another 4 affected newly weaned pigs held in the index farm (Table 1). All of the samples from the asymptomatic control animals were Ni-PoAstV-3 negative.
We determined the complete genomes of 2 Ni-PoAstV-3 strains (NI-Brain/173-2016a/HUN, GenBank accession no. KY073231; and NI-Brain/386-2015/HUN, accession no. KY073232) that originated from 2 affected animals (GD-3 and GD-5) in stage 3 of the disease, chosen at different times (July 2016 and November 2015) of the outbreak (Table 1). These isolates showed 99.5%, 100%, and 98.7%–99.2% aa identities, respectively, to NI-Brain/9-2016a/HUN in the ORF1a, ORF1b, and ORF2 (capsid) regions.
Most of the aa differences between the Ni-PoAstV-3 study strains and the other enteric PoAstV-3 strains are located in the N-terminal part of ORF1a and in the C-terminal part of ORF2 (Table 3). Phylogenetic analysis showed a close relationship between the identified Ni-PoAstV-3 sequences and the known PoAstV-3 strains located within the same larger clade containing most other mamastroviruses with known neurotropic potential (Figure 2).
Detection of Ni-PoAstV-3 in Non-CNS Samples
We detected Ni-PoAstV-3 in multiple non-CNS samples from the respiratory system, lymphoid system, circulatory system, and salivary glands of affected animals (Table 1). We detected virus only in the second PCR round in 1 ileum sample and in 2 of the 3 analyzed fecal samples using nested RT-PCR (Table 1). Samples from internal organs (spleen and kidney) and urine samples tested negative by nested RT-PCR (Table 1).
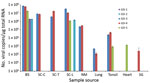
Figure 3. Logarithmic graph of the viral copy numbers of porcine astrovirus type 3 (PoAstV-3) in different organs determined by SYBR Green–based quantitative reverse transcription PCR (RT-qPCR) of samples from 5 symptomatic newly...
We determined the copy number of Ni-PoAstV-3 using SYBR Green–based qPCR. All of the samples that showed nested RT-PCR positivity only in the second (nested) PCR round had negative test results by RT-qPCR, indicating low copy number (<100 copies/µg total RNA) of the virus in that tissue sample. The highest copy number was detected in the brain stem, followed by the spinal cord (Figure 3). Of note, we detected relatively high copy numbers in the tonsil and nasal mucosa samples (Figure 3). The serum of animal GD-3 contained 2.07 × 106 virus copies/mL and of animal GD-4 1.64 × 103 virus copies/mL.
To validate the general presence of Ni-PoAstV-3 in the respiratory system and the absence of the virus in the feces during the acute phase of the illness, we collected additional nasal and anal swab pairs from 5 affected pigs and 13 clinically healthy pigs of the same age (≈25–35 days) from the index farm. Four (80%) of the 5 nasal swab samples from affected animals tested positive but all of the anal swab samples tested negative using nested RT-PCR with primers targeting the RdRp region of Ni-PoAstV-3. The nasal and anal swab samples of the asymptomatic animals were all negative by nested RT-PCR. Because we collected varying amounts of samples by polyester-tipped swabs, we did not perform absolute quantification of Ni-PoAstV-3 by RT-qPCR.
Detection of Ni-PoAstV-3 in Archived FFPE Samples
All but 1 archived FFPE samples from Tázlár and Balmazújváros were positive by nested RT-PCR for Ni-PoAstV-3 using 2 sets of primer pairs targeting the RdRp and capsid genes of Ni-PoAstV-3 (Table 2; Figure 1; Technical Appendix[PDF - 372 KB - 5 pages] Table 2). The spinal cord FFPE sample from Balmazújváros had a negative result using both nested RT-PCR primer sets. The nested RT-PCR positive samples had positive results by ISH (data not shown).
Histology and ISH
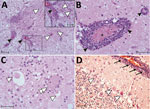
Figure 4. Tissue sections of cervical spinal cord (A), brain stem (B, C) and cerebellum (D) stained with hematoxylin and eosin from a symptomatic newly weaned pig from a farm in Hungary show...

Figure 5. Results of histopathologic testing of central nervous system tissues from 2 symptomatic newly weaned pigs from a farm in Hungary. Sections of the cervical spinal cord (A–E), cerebellum (F–J), and cortex...
Histologically, shared CNS lesions among the animals examined were moderate to marked lymphohistiocytic cell perivascular cuffing with marked vasculitis and neuronal degeneration, necrosis, and neurophagia with multifocal microgliosis and satellitosis (Figure 4). The neuronal necrosis was especially evident in the dorsal and ventral horns of the cervical spinal cord gray matter, although it was also detected in neurons of the Purkinje layer (cerebellum), the medulla oblongata, cerebellar peduncles, and midbrain (Figure 5). Necrotic neurons were variously swollen and hypereosinophilic or shrunken with tinctorial changes including faded, amphophilic, or eosinophilic cytoplasm (Figure 5). Nuclei of affected neurons are pyknotic, karyorrhectic, or losing border definition within the cytoplasm. We performed ISH on 5 affected animals (Table 2). Ni-PoAstV-3 hybridization was predominantly restricted to neurons, including those with visible necrosis and, in the cerebellum in particular, some that were histologically unaffected, although some regions of gliosis (presumed inflammation after neuronal necrosis) also contained viral RNA (Figure 5, panel M). Hybridization was distinct, with punctate to diffuse cytoplasmic staining throughout the cytoplasm. The unique microarchitecture of the Purkinje layer of the cerebellum offered the clear demonstration that viral nucleic acid was present within dendritic processes coursing through the molecular layer (Figure 5, panels G, J). We found no pathologic lesions in other samples from kidneys, liver, gastrointestinal tract, or immune system (data not shown). The samples from the immune system were also negative by Ni-PoAstV-3 ISH (Table 2).
We detected astrovirus RNA in multiple tissues collected during 2015–2017 from newly weaned pigs with encephalomyelitis and posterior paraplegia of unknown origin, with the highest viral load detected in brain stem and spinal cord samples. We detected the same virus in archived brain and spinal cord FFPE samples from similarly affected animals from 2 additional swine herds collected in 2011 and 2014. These data indicate that a genetically similar, neurovirulent astrovirus is circulating in multiple swine farms since 2011 or earlier in Hungary.
According to the refined classification for the assessment of causation (34), the Ni-PoAstV-3 and the observed encephalitis and paraplegia are in a probable causal relationship (Level 2). Paraplegia associated with astrovirus neuroinfection is not unprecedented; minks had astrovirus-induced “shaking mink syndrome” and were reported paraplegic at the final stage of the disease (12,35).
Neurologic signs were observable mainly among newly weaned pigs (Video). The time of weaning, which involves nutritional (from milk to solid feed), social (mixing with different litters without the sow), and environmental (moving to a new pen) changes, is known to be the most stressful period in a pig’s lifetime and is associated with dysfunction of the immune system (36). Furthermore, the inadequate quantity and quality of colostrum intake of sucking piglets, and therefore the presumably low level of specific maternal antibodies due to highly prolific sows with large litters in the index farm, might also contribute to the emergence of the clinical disease. Decreased immune status was frequently present with extraintestinal dissemination of astroviruses in humans and in mice (5,37–41).
Our sequence analyses indicate that the identified astrovirus strains belong to the PoAstV-3 genotype, which clusters within the VA/HMO phylogenetic clade (Figure 2), as do most mammalian strains with known neurotropic potential (6,14,19). However, other canonical human astroviruses outside of the VA/HMO clade could also be associated with CNS disease (41). At the molecular level, the most conspicuous difference between the genomes of neuroinvasive virus and the enteric PoAstV-3 strain U.S.-MO123 is the 27 nt deletions found in the 3′ UTR of the CNS-associated astroviruses. The possible impact of this 3′ UTR deletion on viral tropism is unknown, although neuroinvasive bovine astroviruses also possess 3′ UTR architecture that differs from the diarrhea-associated astroviruses (42).
At the amino acid level, one of the most divergent regions between the neuroinvasive and other PoAstV-3 strains was found at the receptor-interaction domain of ORF2 (Table 3), which contains potential receptor binding sites (43,44). This finding could indicate an altered receptor spectrum and therefore altered tissue tropism of neuroinvasive and enteric PoAstV-3 strains.
PoAstV-3 strains were previously detected only from fecal samples of healthy or diarrheic piglets worldwide (20,22,45). We found that Ni-PoAstV-3 was either undetectable or detected only at low viral loads in the analyzed fecal samples, whereas the virus was generally detectable in the respiratory system of paraplegic pigs. This finding may indicate that CNS infection and replication occur later than enteric replication or that initial replication occurs extraintestinally (e.g., in the respiratory tract). Multiple types of astroviruses were recently identified from nasopharyngeal swabs or lung tissue samples from swine, bovines, and humans with respiratory symptoms including the neurotropic human VA1 strain from a patient with febrile acute respiratory disease (9–11,23), although neither the respiratory tropism nor the airborne transmission of astroviruses has been experimentally confirmed. Therefore, testing of only fecal samples from sick animals may result in underestimation of the incidence of astrovirus in pigs.
We measured the highest viral loads of Ni-PoAstV-3 in brain and spinal cord samples, similar to those found in diseased ovine and human patients with astrovirus-associated encephalitis (5,14). Ni-PoAstV-3 was also detectable in serum specimens and multiple organs of the respiratory, lymphoid, and cardiovascular systems of diseased swine. These results indicate that Ni-PoAstV-3 can result in viremia and disseminated infection involving the brain, spinal cord, and multiple organs during the acute phase of encephalomyelitis and posterior paraplegia. Astroviruses seem to play a role in a common and severe disease (encephalomyelitis and paralysis) in pigs.
The observable histopathologic changes, as well as the neuronal localizations of Ni-PoAstV-3 RNA in CNS samples of paraplegic pigs, are comparable to astrovirus-associated encephalitic cases of minks, humans, and cattle. Similar neuronal degeneration or necrosis with microgliosis in the brain or cerebellum, as well as inflammation of gray matter of the spinal cord, were previously described in cattle with astrovirus-associated nonsuppurative encephalitis (6,35,46,47), which suggests the general course of an astrovirus neuroinfection.
While some astroviruses are known to cause outbreaks of gastroenteritis, astrovirus-associated encephalitis cases have been reported only sporadically among humans, cattle, and sheep (6,14–16,47). The constant presence with recurrent increases of neurologic disease cases in swine farms indicates that natural neuroinvasive astrovirus infections may cause common, severe, persistent epidemics among domestic pigs and constitute an economically important agent threatening livestock and even humans, considering the possible zoonotic and recombinant potential of astroviruses (48).
Our results must be interpreted in the light of some potential limitations, which are currently true for other astrovirus-associated encephalitis studies: the absence of experimental evidence such as in vivo inoculation experiments, which could clarify the true causality between the astrovirus neuroinfection and the manifested CNS symptoms; and the roles of presumed respiratory replication and decreased immune state. Therefore, despite a growing body of scientific data regarding the presence of astroviruses in CNS in different animals, the direct association of astrovirus neuroinfection and encephalomyelitis should be treated with caution. Newly weaned pigs could potentially provide an in vivo animal model to study and clarify this association.
Dr. Boros is a molecular virologist at the Regional Laboratory of Virology, ÁNTSZ Regional Institute of State Public Health Service, Pécs, Hungary. His research interests include virus discovery and viral infectious diseases.
Acknowledgments
We thank Peter Engelmann for help in the cloning experiments.
This work was supported by grants from the Hungarian Scientific Research Fund (OTKA/NKFIH K111615) and Blood System Research Institute. Á.B. and P.P. were supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
References
- Méndez E, Murillo A, Velázquez R, Burnham A, Arias CF. Replication cycle of astroviruses. In: Schultz-Cherry S, editor. Astrovirus research: essential ideas, everyday impacts, future directions. New York: Springer Science + Business Media; 2013. p. 19–47
- Bosch A, Pintó RM, Guix S. Human astroviruses. Clin Microbiol Rev. 2014;27:1048–74. DOIPubMed
- Guix S, Bosch A, Pintó RM. Astrovirus taxonomy. In: Schultz-Cherry S, editor. Astrovirus research: essential ideas, everyday impacts, future directions. New York: Springer Science + Business Media; 2013. p. 97–119.
- Bosch A, Guix S, Krishna NK, Méndez E, Monroe SS, Pantin-Jackwood M, et al. Astroviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. p. 953–9.
- Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis. 2010;16:918–25. DOIPubMed
- Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, et al. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis. 2013;19:1385–92. DOIPubMed
- Meliopoulos V, Schultz-Cherry S. Astrovirus pathogenesis. In: Schultz-Cherry S, editor. Astrovirus research: essential ideas, everyday impacts, future directions. New York: Springer Science + Business Media; 2013. p. 97–119
- Cordey S, Vu DL, Schibler M, L’Huillier AG, Brito F, Docquier M, et al. Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerg Infect Dis. 2016;22:846–53. DOIPubMed
- Padmanabhan A, Hause BM. Detection and characterization of a novel genotype of porcine astrovirus 4 from nasal swabs from pigs with acute respiratory disease. Arch Virol. 2016;161:2575–9. DOIPubMed
- Cordey S, Brito F, Vu DL, Turin L, Kilowoko M, Kyungu E, et al. Astrovirus VA1 identified by next-generation sequencing in a nasopharyngeal specimen of a febrile Tanzanian child with acute respiratory disease of unknown etiology. Emerg Microbes Infect. 2016;5:e67. DOIPubMed
- Ng TFF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol. 2015;89:5340–9. DOIPubMed
- Blomström AL, Widén F, Hammer AS, Belák S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010;48:4392–6. DOIPubMed
- Blomström AL, Ley C, Jacobson M. Astrovirus as a possible cause of congenital tremor type AII in piglets? Acta Vet Scand. 2014;56:82. DOIPubMed
- Pfaff F, Schlottau K, Scholes S, Courtenay A, Hoffmann B, Höper D, et al. A novel astrovirus associated with encephalitis and ganglionitis in domestic sheep. Transbound Emerg Dis. 2017;64:677–82. DOIPubMed
- Vu DL, Bosch A, Pintó RM, Guix S. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses. 2017;9:33. DOIPubMed
- Vu DL, Cordey S, Brito F, Kaiser L. Novel human astroviruses: Novel human diseases? J Clin Virol. 2016;82:56–63. DOIPubMed
- Sato M, Kuroda M, Kasai M, Matsui H, Fukuyama T, Katano H, et al. Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol. 2016;78:66–70. DOIPubMed
- Lum SH, Turner A, Guiver M, Bonney D, Martland T, Davies E, et al. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis. 2016;18:960–4. DOIPubMed
- Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, et al. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis. 2015;60:881–8. DOIPubMed
- Shan T, Li L, Simmonds P, Wang C, Moeser A, Delwart E. The fecal virome of pigs on a high-density farm. J Virol. 2011;85:11697–708. DOIPubMed
- Reuter G, Knowles NJ. Porcine astroviruses. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of swine, 10th ed. Hoboken (NJ): John Wiley & Sons; 2012. p. 487–9.
- Xiao CT, Giménez-Lirola LG, Gerber PF, Jiang YH, Halbur PG, Opriessnig T. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J Gen Virol. 2013;94:570–82. 0
- Xiao CT, Luo Z, Lv SL, Opriessnig T, Li RC, Yu XL. Identification and characterization of multiple porcine astrovirus genotypes in Hunan province, China. Arch Virol. 2017;162:943–52. DOIPubMed
- Ma Z, Lui WO, Fire A, Dadras SS. Profiling and discovery of novel miRNAs from formalin-fixed, paraffin-embedded melanoma and nodal specimens. J Mol Diagn. 2009;11:420–9. DOIPubMed
- Ma Z. Total RNA extraction from formalin-fixed, paraffin-embedded (FFPE) blocks. Bio-protocol. 2012:2:e161 [cited 2017 Sep 20]. http://www.bio-protocol.org/e161
- Boros Á, Pankovics P, Simmonds P, Reuter G. Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS One. 2011;6:e29145. DOIPubMed
- Boros A, Pankovics P, Knowles NJ, Reuter G. Natural interspecies recombinant bovine/porcine enterovirus in sheep. J Gen Virol. 2012;93:1941–51. DOIPubMed
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. DOIPubMed
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMed
- Bouzalas IG, Wüthrich D, Walland J, Drögemüller C, Zurbriggen A, Vandevelde M, et al. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J Clin Microbiol. 2014;52:3318–24. DOIPubMed
- Chu DKW, Poon LLM, Guan Y, Peiris JSM. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–14. DOIPubMed
- Xiao CT, Halbur PG, Opriessnig T. Complete genome sequence of a newly identified porcine astrovirus genotype 3 strain US-MO123. J Virol. 2012;86:13126. DOIPubMed
- Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, et al. Multiple novel astrovirus species in human stool. J Gen Virol. 2009;90:2965–72. DOIPubMed
- Lipkin WI, Anthony SJ. Virus hunting. Virology. 2015;479-480:194–9. DOIPubMed
- Gavier-Widén D, Bröjer C, Dietz HH, Englund L, Hammer AS, Hedlund KO, et al. Investigations into shaking mink syndrome: an encephalomyelitis of unknown cause in farmed mink (Mustela vison) kits in Scandinavia. J Vet Diagn Invest. 2004;16:305–12. DOIPubMed
- Campbell JM, Crenshaw JD, Polo J. The biological stress of early weaned piglets. J Anim Sci Biotechnol. 2013;4:19. DOIPubMed
- Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, Huang HV, et al. Adaptive immunity restricts replication of novel murine astroviruses. J Virol. 2012;86:12262–70. DOIPubMed
- Ng TFF, Kondov NO, Hayashimoto N, Uchida R, Cha Y, Beyer AI, et al. Identification of an astrovirus commonly infecting laboratory mice in the US and Japan. PLoS One. 2013;8:e66937. DOIPubMed
- Farkas T, Fey B, Keller G, Martella V, Egyed L. Molecular detection of novel astroviruses in wild and laboratory mice. Virus Genes. 2012;45:518–25. DOIPubMed
- Marvin SA, Huerta CT, Sharp B, Freiden P, Cline TD, Schultz-Cherry S. Type I interferon response limits astrovirus replication and protects against increased barrier permeability in vitro and in vivo. J Virol. 2015;90:1988–96. DOIPubMed
- Wunderli W, Meerbach A, Güngör T, Berger C, Greiner O, Caduff R, et al. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One. 2011;6:e27483. DOIPubMed
- Bouzalas IG, Wüthrich D, Selimovic-Hamza S, Drögemüller C, Bruggmann R, Seuberlich T. Full-genome based molecular characterization of encephalitis-associated bovine astroviruses. Infect Genet Evol. 2016;44:162–8. DOIPubMed
- Dong J, Dong L, Méndez E, Tao Y. Crystal structure of the human astrovirus capsid spike. Proc Natl Acad Sci U S A. 2011;108:12681–6. DOIPubMed
- Krishna NK. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 2005;18:17–26. DOIPubMed
- Ito M, Kuroda M, Masuda T, Akagami M, Haga K, Tsuchiaka S, et al. Whole genome analysis of porcine astroviruses detected in Japanese pigs reveals genetic diversity and possible intra-genotypic recombination. Infect Genet Evol. 2017;50:38–48. DOIPubMed
- Frémond ML, Pérot P, Muth E, Cros G, Dumarest M, Mahlaoui N, et al. Next-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc. 2015;4:e53–7. DOIPubMed
- Selimovic-Hamza S, Boujon CL, Hilbe M, Oevermann A, Seuberlich T. Frequency and pathological phenotype of bovine astrovirus CH13/NeuroS1 infection in neurologically-diseased cattle: towards assessment of causality. Viruses. 2017;9:12. DOIPubMed
- Ulloa JC, Gutiérrez MF. Genomic analysis of two ORF2 segments of new porcine astrovirus isolates and their close relationship with human astroviruses. Can J Microbiol. 2010;56:569–77. DOIPubMed
![Thumbnail of Phylogenetic analyses of the amino acid sequences of PoAstV-3 isolates (bold) from 3 symptomatic newly weaned pigs (GD-1, GD-3, and GD-5; see Table 1) from a farm in Hungary compared with reference isolates. A) ORF1a; B) ORF1b; C) ORF2. We included in the analysis available strains of the closest relatives (identified by blastx search [https://blast.ncbi.nlm.nih.gov/Blast.cgi]) of neuroinvasive PoAstV-3, all of the known porcine astroviruses with available complete coding sequences,](https://wwwnc.cdc.gov/eid/images/17-0804-F2-tn.jpg)






















.png)











No hay comentarios:
Publicar un comentario