Notes from the Field: Investigation of an Outbreak of Salmonella Paratyphi B Variant L(+) tartrate + (Java) Associated with Ball Python Exposure — United States, 2017
Weekly / May 19, 2018 / 67(19);562–563
Vikram Krishnasamy, MD1,2; Lauren Stevenson, MHS2,3; Lia Koski, MPH2,4; Marilee Kellis, MPH5; Betsy Schroeder, DVM6; Madhura Sundararajan, MPH6; Stephen Ladd-Wilson, MS7; Ashley Sampsel7; Mike Mannell, MPH8; Andrew Classon2; Darlene Wagner, PhD2,9; Kelley Hise, MPH2; Heather Carleton, PhD2; Eija Trees, DVM, PhD2; Linda Schlater, DVM10; Kristina Lantz, DVM10; Megin Nichols, DVM2 (View author affiliations)
View suggested citationIn July 2017, PulseNet, the national molecular subtyping network for foodborne disease surveillance, identified a cluster of five SalmonellaParatyphi B variant L(+) tartrate + (Java) clinical isolates that were indistinguishable by pulsed-field gel electrophoresis (PFGE). Initial questionnaires administered by state and local health department investigators indicated animal exposure as a possible source of infection, with all five patients reporting snake exposure. An outbreak investigation was initiated to identify the source of infection.
A case was defined as isolation of Salmonella Paratyphi B variant L(+) tartrate + (Java) from June 17, 2017, to July 23, 2017, with a PFGE enzyme pattern indistinguishable from the outbreak strain. A snake-specific questionnaire regarding snake type, snake purchase location, and reptile food, including feeder rodents, was developed and administered to patients by state and local health department investigators. In addition, animal and environmental sampling was conducted at patient residences. Traceback of patients’ snakes was conducted by contacting snake purchase locations to identify common suppliers. Finally, whole genome sequencing (WGS) was performed on clinical, environmental, animal, and pet food isolates to further characterize their genetic relatedness, measured in single nucleotide polymorphism (SNP) differences (1).
Five cases were identified in four states: one each in Arizona, Oklahoma, and Oregon, and two in Indiana from different households with no epidemiologic link. Median patient age was 10 years (range = <1–40 years), and four were female. No patient was hospitalized, and no deaths occurred. Five patients or their proxies completed the snake-specific questionnaire, four of whom reported exposure to a ball python in the residence. Ball python sampling occurred in the Arizona, Oregon, and one of the Indiana patient residences by sampling the python cloaca, environment, water, and bedding. Feeder rodent sampling occurred in the Arizona and Indiana patient residences. No common suppliers of either ball pythons or feeder rodents were identified by traceback.
A ball python bedding sample from the Arizona residence and a ball python cloacal sample from the Indiana residence yielded Salmonella Paratyphi B variant L(+) tartrate + (Java). Sampling also identified Salmonella Mbandaka from a feeder rodent and a ball python at the Arizona residence. In addition, Oregon ball python environmental samples yielded Salmonella Oranienburg. Further investigation of the Salmonella Mbandaka and Oranienburg isolates did not identify human illnesses linked to snake exposure. Finally, the National Veterinary Services Laboratories (NVSL) identified three Salmonella Paratyphi B var L(+) tartrate + (Java) isolates from other pythons in 2017.
WGS analysis indicated that among human isolates, only the two Indiana patient isolates were closely related genetically (0–2 SNP differences) (Figure). The Arizona, Oklahoma, and Oregon patient isolates were not closely related genetically to each other or to the Indiana patient isolates. In addition, the Arizona ball python bedding sample was indistinguishable from the Arizona patient isolate (0 SNP differences), and the Indiana ball python and environmental samples were closely related to both Indiana patient isolates (0–2 SNP differences). Finally, the three python isolates identified at NVSL were not closely related to any human, animal, or environmental isolate.
Python regius, also known as ball or royal pythons, are a python species native to sub-Saharan Africa. Their tame nature and small size relative to other pythons make them popular pets in the United States (2,3). However, like other reptiles, ball pythons are known carriers of multiple Salmonella serovars (4). As a result, CDC recommends that children aged <5 years avoid contact with reptiles (5). The median age of patients in this investigation was 10 years, indicating that children aged >5 years are also at risk for illness. Identifying and investigating zoonotic clusters of salmonellosis require a One Health (6) approach using multidisciplinary collaboration with departments of agriculture and health.
Acknowledgments
Hayley Yaglom, Joli Weiss, Kaitlyn Snyder, Ginny De La Cruz, Roumen Penev, Joe Huynh, Nannette Washington, Logan Fink, Joel Sevinsky.
Conflict of Interest
No conflicts of interest were reported.
Corresponding author: Vikram Krishnasamy, vkrishnasamy@cdc.gov, 404-718-5504.
1Epidemic Intelligence Service, CDC; 2Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, CDC; 3CAITTA, Inc., Herndon, Virginia; 4Oak Ridge Institute for Science and Education, Oak Ridge Tennessee; 5Arizona Department of Health Services; 6Indiana State Department of Health; 7Oregon Health Authority, Public Health Division; 8Oklahoma State Department of Health; 9IHRC, Inc., Atlanta, Georgia; 10National Veterinary Services Laboratories, Science, Technology and Analysis Services, Veterinary Services, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Washington, DC.
References
- CDC. Whole genome sequencing (WGS). Atlanta, GA: US Department of Health and Human Services, CDC; 2017. https://www.cdc.gov/ncezid/dfwed/keyprograms/tracking-foodborne-illness-wgs.html
- Reed RN. An ecological risk assessment of nonnative boas and pythons as potentially invasive species in the United States. Risk Anal 2005;25:753–66. CrossRef PubMed
- De Vosjoli PD, Barker T, Klingenberg R. The ball python manual. Los Angeles, CA: i5 Publishing; 2012.
- Ebani VV, Cerri D, Fratini F, Meille N, Valentini P, Andreani E. Salmonella enterica isolates from faeces of domestic reptiles and a study of their antimicrobial in vitro sensitivity. Res Vet Sci 2005;78:117–21. CrossRef PubMed
- CDC. Healthy pets, healthy people—infants and young children. Atlanta, GA: US Department of Health and Human Services, CDC; 2017. https://www.cdc.gov/healthypets/specific-groups/children.html
- CDC. One health. Atlanta, GA: US Department of Health and Human Services, CDC; 2018. https://www.cdc.gov/onehealth/index.html
 FIGURE. Whole-genome sequencing analysis of isolate genomes of Salmonella Paratyphi B variant L(+) tartrate + (Java) from human, from Python regius, and from environmental sources associated with the outbreak investigation — United States, 2017*
FIGURE. Whole-genome sequencing analysis of isolate genomes of Salmonella Paratyphi B variant L(+) tartrate + (Java) from human, from Python regius, and from environmental sources associated with the outbreak investigation — United States, 2017*
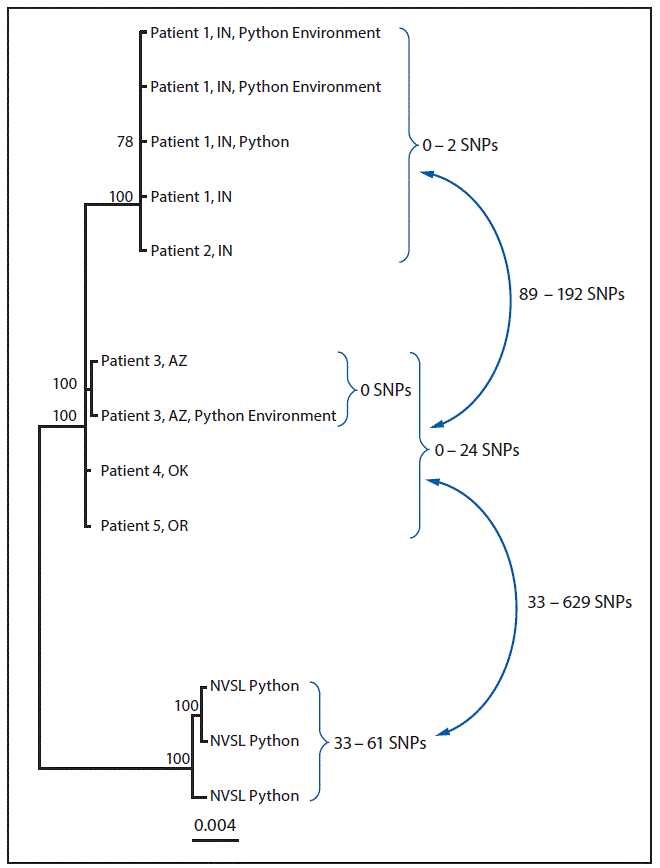
Abbreviations: AZ = Arizona; hqSNP = high-quality single nucleotide polymorphism; IN = Indiana; NVSL = National Veterinary Services Laboratories; OK = Oklahoma; SNPs = single nucleotide polymorphisms.
* Analysis performed using Lyve-SET version 1.1.4f (https://github.com/lskatz/lyve-SET) and hqSNPs were called at >20x coverage, >95% read support, and allowed Flanking set to five base pairs. Reference used was draft assembly of Arizona case (78 contigs) without phage masking.
† Three isolates from different pythons submitted to NVSL for testing.
The figure above is a diagram showing a whole-genome sequencing analysis of isolate genomes of Salmonella Paratyphi B variant L(+) tartrate + (Java) from human, from Python regius, and from environmental sources associated with the outbreak investigation in the United States in 2017.






















.png)












No hay comentarios:
Publicar un comentario