
Volume 25, Number 3—March 2019
Dispatch
In Vivo Selection of a Unique Tandem Repeat Mediated Azole Resistance Mechanism (TR120) in Aspergillus fumigatus cyp51A, Denmark
On This Page
Maiken C. Arendrup , Jan B. Gertsen, Karen M.T. Astvad, Kristine B. Degn, Anders Løkke, Marc Stegger, Paal S. Andersen, and Lise Kristensen
, Jan B. Gertsen, Karen M.T. Astvad, Kristine B. Degn, Anders Løkke, Marc Stegger, Paal S. Andersen, and Lise Kristensen
Abstract
We report a fatal aspergillosis case in which STRAf typing and whole-genome sequencing substantiated in vivo emergence of an azole-resistant Aspergillus fumigatus with a 120-bp tandem repeat in the promoter region of cyp51A. This event, previously restricted to the environment, challenges current understanding of azole resistance development in A. fumigatus.
Azole antifungal drug resistance in Aspergillus fumigatus is a concern for patients with aspergillosis because of increased risk for disease and death (1). Two routes of acquiring azole resistance have been identified: 1) in vivo, as a consequence of long-term azole treatment; and 2) ex vivo, in the environment, resulting from the use of azole fungicides in crop protection. The underlying mechanisms are primarily linked to structural changes or upregulation of the azole target lanosterol 14 α-demethylase encoded by cyp51A (1). Most environmentally induced resistance mechanisms involve tandem repeats (TRs) in the promoter region of cyp51A coupled with nonsynonymous mutations, TR34/L98H and TR46/Y121F/T289A (1). However, in vivo resistance development has primarily been associated with nonsynonymous mutations in cyp51A-inducing amino acid substitutions of hot spots (e.g., G54, G138, M220, and G448) or non–cyp51A-mediated mechanisms, but not a tandem repeat (1). We describe a clinical case of infection with azole-resistant A. fumigatus that acquired a 120-bp tandem repeat (TR120) resistance mechanism during long-term azole treatment. The finding was substantiated by whole-genome sequencing (WGS).
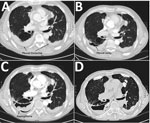
Figure 1. Sequential thoracic computed tomography scan images illustrating the gradual progression from pleural thickening to cavity formation and development of an aspergilloma in a patient with Aspergillus fumigatusinfection, Denmark, 2013. A)...
In 2013, a 69-year-old man who was a former smoker with chronic obstructive pulmonary disease (COPD) and severe airflow obstruction sought care at the University Hospital in Århus, Denmark, because of gradually worsening dyspnea, cough, and expectoration. Previously, in 2011, imaging (Figure 1, panel A) and 2 thoracoscopies had been conducted because of suspicion of malignant mesothelioma. Further histopathologic examination and cultures revealed inflammation but no malignancy or mold infection. Subsequently, in 2012, a fistula between pleura and skin led to a persistent air-containing pleural cavity in the right side (Figure 1, panel B). In 2014, a fungus ball in the pleural cavity was found (Figure 1, panel C). Aspergillus IgG titer was 1:25,600 (reference range <1:200), and azole-susceptible A. fumigatus was cultured from sputum (P-1, May 2014). Voriconazole (200 mg 2×/d) was given, alternating with posaconazole (300 mg/d) for 2 years until clinical failure, and 2 azole-resistant A. fumigatus isolates were cultured from a new sputum sample (P-2 and P-3, June 2016). Despite amphotericin B inhalations followed by liposomal amphotericin B (3 mg/kg 1×/d), the patient died because of severe hemoptysis 1 year later in 2017.
Three A. fumigatus patient isolates (P-1, P-2, and P-3) were available for confirmatory species verification, reference susceptibility testing defined by the European Committee on Antimicrobial Susceptibility Testing using protocol for molds (E.Def 9.3), cyp51A Sanger sequencing (using wild-type reference sequence AF338659), and genotyping using the short tandem-repeat Aspergillus fumigatus (STRAf) assay (2,3) (Table). We included 4 A. fumigatusisolates representing relevant cyp51A profiles as control strains (SSI-3614 [wild-type], SSI-7828 [TR34/L98H], SSI-5717 [TR46/Y121F/T289A], and SSI-5197 [F46Y/M172V/E427K]). We detected 3 common Cyp51A variants (F46Y, M172V, and E427K) in the susceptible patient isolate P-1 (GenBank accession no. MG972984). Pan-azole resistance was observed for P-2 and P-3, and both shared cyp51A profiles with P-1 but also harbored a TR120mechanism (GenBank accession no. MG972983) in the promoter region (Table). All patient isolates had identical STRAf genotypes suggesting that they were isogenic (Table ) (4). Furthermore, the STRAf profile was unique among A. fumigatus isolates genotyped in Denmark (Appendix Figure).

Figure 2. Unrooted phylogenetic tree based on whole-genome sequencing of 2 patient isolates (P-1 and P-3) and 5 reference strains to highlight relatedness between Aspergillus fumigatus isolates, Denmark, 2018. We inferred relatedness by...
We performed WGS for P-1, P-3, and all control strains to investigate relatedness and other potential mechanisms conferring azole resistance. We subjected total DNA (≈10 ng/µL) to WGS (NextSeq 550; Illumina, https://www.illumina.com) by using Nextera DNA library preparation kit (Illumina) and following the manufacturer’s instructions. We used NASP (5) to detect single-nucleotide polymorphisms (SNPs) after removal of duplicated regions in the A. fumigatus strain Af293 chromosomes (http://www.aspergillusgenome.org, genome version s03-m05-r09) using NUCmer (6). We inferred relatedness by using FastTree version 2.1.5 (7) and a 77.69% core genome (Table; Figure 2). To increase resolution, we conducted a subanalysis for P-1 and P-3 (core genome 79.71%), which identified 41 SNP differences; 6 of the SNPs were nonsynonymous in genes with no previous reported association to azole resistance (Appendix Table 1), and 35 were either synonymous or in noncoding regions (Appendix Table 2).
WGS revealed 41 SNP differences between the susceptible and the resistant patient A. fumigatus isolates that evolved during 2 years, similar to a previously described case of in-host microevolution of A. fumigatus (4). This finding substantiated an isogenic relationship between P-1 and P-3 and demonstrated that the TR120 resistance mechanism emerged from P-1, probably during long-term azole therapy. Furthermore, WGS results supported the conclusion that the TR120 was the sole mechanism of azole resistance in the azole-resistant patient isolates.
To our knowledge, the TR120 is a novel azole-resistance mechanism in A. fumigatus, and the in vivo selection of a tandem repeat in the promoter of cyp51Ais unique. The de novo acquisition of a TR has not previously been shown in vitro or in the environment (i.e., no isolates with L98H or Y121F+T289A combined with wild-type promoters have been reported). However, triplication of an existing TR34 on tebuconazole exposure was selected in vitro, and a novel variant, TR463, found in clinical and environmental samples, has been derived from sexual mating between TR46 parents (8,9).
Azole resistance involving TRs in the promoter region has been associated exclusively with environmental fungicide selection pressure in A. fumigatus and other plant pathogens. Furthermore, although asexual propagation of A. fumigatus with TR34/L98H or TR46/Y121F/T289A resistance mechanisms is widespread in the environment, the extent of de novo selection of TR34/L98H and TR46/Y121F/T289A is unclear (10). One hypothesis describes both environmental resistance mechanisms as being derived from single events of sexual reproduction (in environmental habitats) combining the TR with a cyp51A mutant. In addition, sexual reproduction might have led to a high genetic diversity among environmental azole-resistant A. fumigatus, which otherwise might have indicated multiple origins (10). Our finding might challenge the perception that TR azole-resistance mechanisms are exclusive to the environment and might warrant the question of whether TR34/L98H and TR46/Y121F/T289A derive from single events.
Hypothetically, the patient might initially have inhaled isogenic isolates with and without TR120, the resistant one being undetected. However, a patient being co-infected de novo by a susceptible and an isogenic resistant strain has not been previously reported and is considered highly unlikely.
Long-term and subtherapeutic antifungal treatment might facilitate selection of resistance (11). Therapeutic drug monitoring was performed once in this patient but without information if the sample was taken according to guidelines as a trough level (lowest level after dosage). Thus, despite a concentration of 4.3 mg/L (within the recommended trough range), potential subtherapeutic levels during the 200 mg 2×/d dosing scheme cannot be ruled out. The F46Y/M172V/E427K substitutions in Cyp51A, found in both susceptible and resistant isolates, have been suggested to play no role or only a minor role in reduced azole susceptibilities (12,13). TRs in the promoter region of cyp51A have previously been linked to increased cyp51A gene expression and MICs because of duplicated srbA transcription factor binding motifs (SRE1 and SRE2) leading to increased expression of cyp51A (14,15). Taken together, our data suggest that TR120 alone is an important driver of pan-azole resistance at a level comparable to that known to be mediated by the TR34/L98H mechanism.
Our WGS results might obviate the desire for in vitro experiments testing the TR120 mechanism in laboratory-engineered mutants. Further dissection of the WGS data can help elucidate potential genetic drivers of TR acquisition and add further knowledge as to whether the TR34/L98H and TR46/Y121F/T289A resistance genotypes derived from a single origin. This report adds another piece to the complex picture of emerging azole-resistant A. fumigatus and might serve to stimulate further research.
Dr. Hare is a molecular biologist at the Mycology Laboratory at Statens Serum Institut, Copenhagen, Denmark, where he completed his PhD on antifungal drug resistance in 2016. Besides antifungal resistance, his main research interests are molecular fungal diagnostics.
Acknowledgment
We thank the entire Mycology Laboratory at Statens Serum Institut for their invaluable work in culturing, identification, and antimicrobial susceptibility testing. We acknowledge the staff responsible for the WGS workflow for their expedited service. We also thank laboratory technicians involved in PCR and DNA analyses, including Nissrine Abou-Chakra, for their invaluable aid and expertise in molecular biology.
References
- Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing. EUCAST Technical Note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Clin Microbiol Infect. 2008;14:982–4. DOIPubMed
- Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom. 2016;2:e000074.
- Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 2003;00:10.3.1–10.3.18.
- Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, et al. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. MBio. 2015;6:e00536.PubMed
Figures
Table
Cite This ArticleOriginal Publication Date: 1/31/2019





















.png)












No hay comentarios:
Publicar un comentario