
Creative Minds: Programming Cells to Write Their Own Memoirs
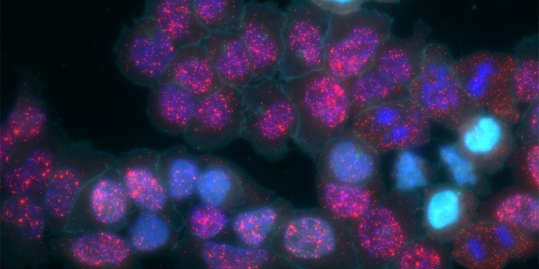
Caption: MEMOIR cells variably activated (cyan). The recorded information is then read out to visualize certain RNA transcripts (red).
Credit: Elowitz and Cai Labs/Caltech
Credit: Elowitz and Cai Labs/Caltech
One of the most fascinating challenges in biology is understanding how a single cell divides and differentiates to form a complex, multicellular organism. Scientists can learn a lot about this process by tracking time-lapse images through a microscope. But gazing through a lens has its limitations, especially in the brain and other opaque and inaccessible tissues and organs.
With support from a 2017 NIH Director’s Transformative Research Program, a California Institute of Technology (Caltech) team now has a way around this problem. Rather than watching or digging information out of cells, the team has learned how to program cells to write their own molecular memoirs. These cells store the information right in their own genomic hard drives. Even better, that information is barcoded, allowing researchers to read it out of the cells without dissecting tissue. The programming can be performed in many different cell types, including stem or adult cells in tissues throughout the body.
Using this approach, dubbed MEMOIR (short for Memory by Engineered Mutagenesis with Optical In-Situ Readout), the researchers are now able to read telling details from a cell’s past. These details include determining its cellular ancestors, the cells it “talks” to. and even when such “conversations” took place.
As described recently in the journal Nature [1], the team—including Michael Elowitz, Long Cai, and Carlos Lois—developed MEMOIR using two innovative tools. The first is the genome-editing system known as CRISPR/Cas9. The second is a microscopy technique developed in the Cai lab called seqFISH [2], which makes it possible to tag and visualize the fluorescence of hundreds of specific RNA transcripts inside single cells.
To program MEMOIR into an individual cell, Kirsten Frieda, Sahand Hormoz, James Linton, and other members of the team insert many copies of an identical DNA sequence. Each individual sequence, which they call a “scratchpad,” is attached to another uniquely identifying barcoded sequence. The Cas9 enzyme, often likened to a pair of molecular scissors, cuts away those scratchpads at random over time. When a scratchpad is removed, the barcode remains to be passed on to its cellular progeny, revealing the history of that event.
To visualize the scratchpads and their associated barcodes, the researchers use a set of distinct molecular, seqFISH probes. By observing the patterns of scratchpad loss, the researchers can make accurate inferences about how a particular cell is related to all of the other cells.
The researchers say these programmed cells can also record particular molecular signals or other events of interest. For example, cells could keep a record of changes in gene expression that occurred during important cell fate transitions. Each time a target signal is received, the cell would respond by making a particular mutation in a specific, non-coding portion of the genome. Those mutations can then be read out later using seqFISH.
The researchers have already provided their proof-of-principle that MEMOIR works when programmed into the embryonic stem cells of mice [1]. By inserting multiple MEMOIR systems into cells in parallel, they are now engineering cells to record multiple types of information at the same time.
The researchers envision that MEMOIR will allow them to tackle big questions in more complex tissues. One of their first targets is the brain. They say MEMOIR can help to resolve fundamental and controversial questions—for instance, whether the six layers found in the cerebral cortex, the outermost part of the brain, arise from a single progenitor cell or several distinct progenitors.
Ultimately, they hope to apply MEMOIR to tracking cells responsible for a tumor’s spread from one part of the body to another. There’s still plenty of programming work to be done. But the days of reading cellular memoirs are coming.
References:
[1] Synthetic recording and in situ readout of lineage information in single cells. Frieda KL, Linton JM, Hormoz S, Choi J, Chow KK, Singer ZS, Budde MW, Elowitz MB, Cai L. Nature. 2017 Jan 5;541(7635):107-111.
[2] In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Shah S, Lubeck E, Zhou W, Cai L. Neuron. 2016 Oct 19;92(2):342-357.
Links:
Elowitz Lab (Caltech)
Cai Lab (Caltech)
Lois Lab (Caltech)
MEMOIR Project Information (NIH RePORTER)
NIH Director’s Transformative Research Program (Common Fund)
NIH Support: National Institute of Mental Health; Common Fund





















.jpg)












No hay comentarios:
Publicar un comentario