
Volume 23, Number 8—August 2017
Research
Clinical Laboratory Values as Early Indicators of Ebola Virus Infection in Nonhuman Primates
On This Page
Ronald B. Reisler, Chenggang Yu, Michael J. Donofrio, Travis K. Warren, Jay B. Wells, Kelly S. Stuthman, Nicole L. Garza, Sean A. Vantongeren, Ginger C. Donnelly, Christopher D. Kane, Mark G. Kortepeter, Sina Bavari, and Anthony P. Cardile
Abstract
The Ebola virus (EBOV) outbreak in West Africa during 2013–2016 demonstrated the need to improve Ebola virus disease (EVD) diagnostics and standards of care. This retrospective study compared laboratory values and clinical features of 3 nonhuman primate models of lethal EVD to assess associations with improved survival time. In addition, the study identified laboratory values useful as predictors of survival, surrogates for EBOV viral loads, and triggers for initiation of therapeutic interventions in these nonhuman primate models. Furthermore, the data support that, in nonhuman primates, the Makona strain of EBOV may be less virulent than the Kikwit strain of EBOV. The applicability of these findings as potential diagnostic and management tools for EVD in humans warrants further investigation.
The Ebola virus (EBOV) outbreak in West Africa during 2013–2016 highlighted the need to improve Ebola virus disease (EVD) diagnostics and standards of care (1). With regard to standards of care and EVD outcomes, it is important to explore potential factors associated with improved survival. In previous epidemics, dating back to 1976, EVD case fatality rates ranged from 47% to 90% (2). The Centers for Disease Control and Prevention (CDC) reported a crude death rate of 40% (11,310 deaths/28,616 cases) when including suspected, probable, and confirmed cases from the West Africa outbreak; however, when only confirmed cases were included, the EVD case fatality rate was 74% (11,310 deaths/15,227 cases), consistent with historical rates (1).
Analysis of the most recent EBOV epidemic and previous outbreaks identified predictors associated with decreased survival: high quantitative viral load (3–7); low PCR cycle threshold (8–12); age (very young and very old) (5,6,10,13–16); male sex (12,17); country of residence (1,17); levels of D-dimer (18), aspartate aminotransferase (AST) (5,10,11), blood urea nitrogen (BUN) (5), and serum creatinine (5,10); and clinical symptoms of diarrhea, pain, myalgia, hemorrhage, and difficulty breathing (10,12,15,19,20). Of the various EVD animal models that have been developed, those using nonhuman primates (NHPs) appear to most closely reproduce the known features of lethal disease in humans. Herein, we summarize and compare the clinical features and laboratory values of 3 NHP lethal models of EVD and explore features associated with early manifestations of infection and improved survival. Similar to Janvier et al. (11), who recommended the use of high AST levels in humans as a surrogate marker of EBOV viral load and, therefore, disease detection and survival, we explored whether NHP laboratory data would lend support to the use of clinical laboratory values as predictors of survival and surrogates for EBOV viral loads. Further analysis was conducted to determine if clinical laboratory values could be used for indication of infection with the goal of developing a standardized trigger for the initiation of treatment in the NHP model.
Animal Use and Viral Challenge
The 30 NHPs described herein served as control animals in larger therapeutic studies conducted in 2014 and 2015. We retrospectively analyzed existing data from those studies. The NHP experiments and tests for this study were performed by the same researchers at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID; Fort Detrick, Frederick, MD, USA), using the same institutional standard operating procedures and processes, the same laboratory instruments, and the same standardized EBOV challenge (1,000 PFU administered intramuscularly [IM]).
We collected blood samples from all 30 NHPs immediately before virus challenge (day 0) and at 3, 5, and 7 days postinoculation (dpi). Methods for EBOV challenge of the NHPs have been described in detail (21). In brief, we first IM inoculated 18 rhesus macaques (Macaca mulatta; 3 groups of 6 animals, male and female, weighing 3.71–7.26 kg) with a target titer of 1,000 PFU of the Kikwit strain of EBOV (EBOV Kikwit; back titration titer range 950–1,358 PFU). Next, we IM inoculated 6 cynomolgus macaques (M. fascicularis; male and female, weighing 3.49–7.33 kg) with a target titer of 1,000 PFU of EBOV Kikwit (back titration titer 1,600 PFU). The EBOV Kikwit strain we used is a USAMRIID stock virus, EBOV H.sapiens-tc/COD/1995/Kikwit-9510621; this virus was primarily the 7U (7 uridylyls) variant at the mRNA editing site. This challenge virus was propagated from a clinical specimen by using cultured cells for a total of 4 passages. Last, we IM inoculated 6 rhesus macaques (male and female, weighing 4.79–5.40 kg) with a target titer of 1,000 PFU (back titration titer 800 PFU) of the USAMRIID stock virus EBOV H.sapiens-tc/LBR/2014/Makona (EBOV Makona); this virus was also primarily the 7U variant at the mRNA editing site. This challenge virus was propagated from a clinical specimen by using cultured cells for a total of 2 passages.
We conducted all studies in Biosafety Level 4 containment. Beginning on day 0 and continuing for the duration of the in-life phase, we recorded clinical observations and closely monitored animals at least 3 times daily for disease progression (22). According to protocol, we provided the NHPs with basic support with regard to pain, oral hydration, and antimicrobial drugs. We administered antimicrobial drugs only if the facility veterinarian diagnosed a secondary bacterial infection. Moribund animals were euthanized on the basis of prespecified criteria (22).
Clinical Laboratory Samples
When possible, we processed and analyzed samples obtained for analyses within 6 h of collection. We used a Vitros 350 Chemistry System (Ortho Clinical Diagnostics, Raritan, NJ, USA) to analyze serum chemistries; an Advia 120 Hematology Analyzer (Siemens, Tarrytown, NY, USA) with multispecies software to analyze hematology parameters; and a Sysmex CA-1500 (Siemens) for coagulation analyses. The samples yielded a panel of 46 routine clinical laboratory values (21).
Viral RNA
We used quantitative real-time PCR (qRT-PCR) to determine viral RNA copy numbers in plasma samples collected at prespecified time points (21). No definition has been established for high viral load in this qRT-PCR assay or in NHP models of EVD. Thus, we used a value of 9 log10 RNA copies/mL as a cutoff value for high versus low viral load. The rationale for this cutoff was that the median viremia value for the EBOV Kikwit–infected macaques at 5 dpi and that for the EBOV Makona–infected macaques at 7 dpi was ≈9 log10 copies/mL.
Statistical Analysis
We performed univariate and multivariate regression modeling of available demographic and laboratory data and used Mann-Whitney U test and Fischer exact test, where appropriate, for the descriptive analyses. To assess the effect of variables on survival time (measured in hours), we used linear and logistic regression models with Microsoft Excel (Microsoft Corp, Redmond, WA, USA) and Stata 12 (StataCorp LLC, College Station, TX, USA). We used p value thresholds of <0.05 for statistical tests and included an adjusted p value of ≤0.001, based on a simplified Bonferroni correction, for multiple comparisons (Technical Appendix[PDF - 245 KB - 4 pages] Table 1).
We used the receiver operating characteristic (ROC) curve to illustrate a predictor’s performance in 2 metrics, with 1 as a tradeoff of another (e.g., sensitivity and specificity); we obtained the ROC curve by varying the laboratory threshold values of a tested predictor. Using ROC area under the curve (AUC), we evaluated the ability of routine laboratory values (alone or in combination) and log10 plasma viral load RNA concentrations obtained at 3, 5, or 7 dpi to predict infection. The combinations of routine laboratory values analyzed were chosen on the basis of the characteristics of individual laboratory values, clinical relevance, and likely access to the test in a field Ebola treatment unit. Day 0 laboratory values were used as baseline values. Because blood samples for viral RNA assessment and routine laboratory analysis were obtained before virus challenge, day 0 values were used to represent uninfected animals. Values obtained at 3, 5, and 7 dpi were used to represent infected animals because all NHPs were experimentally IM inoculated with EBOV (1,000 PFU target dose), and plasma viral RNA was detected in a sample from at least 1 sampling event for all NHPs included in this analysis. As part of this analysis, we combined the following variables: AST; lactate dehydrogenase (LDH); C-reactive protein (CRP); and the combination of AST, LDH, CRP, and hemoglobin (Hgb). A combined variable C was defined as the mean value of all normalized variables and vi:
.
vi was normalized by dividing the sum of its mean and standard deviation of baseline values (i.e., values on day 0). si is 1 (or −1) for variables whose value increases (or decreases) after infection. m is the number of independent variables to be combined. Values of variable C were calculated for each sample on each day (including day 0) after μi, σi, and si were determined. We denote the 2 combined variables as (AST+LDH+CRP) and (AST+LDH+CRP−Hgb) because AST, LDH, and CRP values increase after infection, whereas Hgb decreases after infection.Ethics Statement
Animal research at USAMRIID was conducted under an Institutional Animal Care and Use Committee–approved protocol in compliance with the Animal Welfare Act, US Public Health Service policy, and other federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
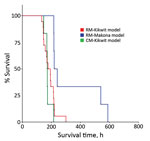
Figure 1. Kaplan–Meier survival analysis for each of 3 nonhuman primate models of Ebola virus disease: rhesus macaque model with EBOV Kikwit strain (n = 18 monkeys); rhesus macaque model with EBOV Makona...

Figure 2. log10 RNA level, by day after EBOV challenge, for each of 3 nonhuman primate models of Ebola virus disease. Box and whisker plots were created by using the available data for...
Similarities and differences in baseline summary characteristics of the 3 groups of NHPs are worth noting (Tables 1, 2) as well as similarities and differences in Kaplan–Meier survival analysis (Figure 1). Median RNA viral loads peaked in all 3 NHP groups at 7 dpi (Figure 2), but RNA viral loads for some EBOV Kikwit–infected NHPs peaked at 5 dpi. At each time point, viremia values ranged widely between the 3 groups. The only significant differences between the NHP groups were at 3 dpi, when the EBOV Kikwit–infected rhesus macaques had higher mean log10RNA values (4.50 RNA copies/mL [range <3.0 to 6.54]) than the EBOV Makona–infected rhesus macaques, all of whom had log10 RNA values below the limit of detection (<3.00 RNA copies/mL; p<0.001), and at 5 dpi, when the EBOV Kikwit–infected rhesus macaques had higher mean log10 RNA values (8.94 RNA copies/mL [range 5.94 to 10.47]) than the EBOV Makona–infected rhesus macaques (6.57 RNA copies/mL [range <3.0 to 9.33]; p<0.049).

Figure 3. Survival curves, stratified by high (>9 log10 RNA copies/mL) and low (<9 log10 RNA copies/mL) viral loads, for each of 3 nonhuman primate models of Ebola virus disease. A) Comparison of...
In the EBOV Kikwit–infected rhesus macaques, the median survival time was significantly different between animals with high viral loads (214.6 h) and those with low viral loads (148.0 h) (p = 0.013) (Figure 3). In general, viral load correlated with survival time as early as 3 dpi for EBOV Kikwit–infected rhesus macaques (r = 0.57; p = 0.013); 5 dpi for EBOV Kikwit–infected cynomolgus macaques (r = 0.75; p = 0.084); and 7 dpi for EBOV Makona–infected rhesus macaques (r = 0.90; p = 0.016).
EBOV Kikwit–Infected Rhesus Macaques versus EBOV Makona–Infected Rhesus Macaques
Among rhesus macaques, survival time was longer for those infected with EBOV Makona (337.5 hours) than those infected with EBOV Kikwit (186.9 hours; p = 0.005) (Table 1). Clinical assessments showed significant differences between clinical disease progression in EBOV Kikwit–infected and EBOV Makona–infected NHPs at 5 and 7 dpi (Table 1). In addition, we found significant differences in laboratory assessments at 5 and 7 dpi (Table 2). And, at 5 dpi, log10 plasma concentrations of viral RNA were significantly higher among EBOV Kikwit–infected rhesus macaques (8.94 RNA copies/mL) than among EBOV Makona–infected rhesus macaques (6.57 RNA copies/mL; p = 0.049).
EBOV Kikwit–Infected Rhesus Macaques versus EBOV Kikwit–Infected Cynomolgus Macaques
We found no significant difference in survival time between EBOV Kikwit–infected rhesus (186.9 hours) and cynomolgus (175.2 hours) macaques. In addition, at 5 and 7 dpi, we found no significant difference in clinical findings between these 2 groups (Table 1). However, there were subtle differences in laboratory values (Table 2). Compared with rhesus macaques, cynomolgus macaques had worsened markers of renal function at 7 dpi, as evidenced by mean laboratory values: BUN levels of 112.6 mg/dL for cynomolgus macaques versus 58.7 mg/dL for rhesus macaques (p = 0.015); serum creatinine levels of 24.3 mg/dL for cynomolgus macaques versus 2.3 mg/dL for rhesus macaques (p = 0.037); and serum potassium levels of 6.2 mEq/L for cynomolgus macaques versus 4.2 mEq/L for rhesus macaques. Furthermore, at 7 dpi, mean platelet counts tended to be lower for rhesus (89.6 × 103/mm3) than cynomolgus (142.4 × 103/mm3; p = 0.047) macaques (Table 2).
Regression Analyses for Predicting Viral Load from Routine Laboratory Values
Due to small sample sizes, we limited our presentation of regression analyses to the 18 EBOV Kikwit–infected rhesus macaques. In a univariate regression model, log10 plasma concentrations of viral RNA correlated significantly with survival time at peak viremia (5–7 dpi) and at 3, 5, and 7 dpi (Technical Appendix[PDF - 245 KB - 4 pages]). At 5 dpi, the following laboratory values correlated significantly with time to death and with plasma viral load: platelet counts; prothrombin time; and levels of AST; alanine aminotransferase (ALT); LDH; and creatine phosphokinase (CPK) (Technical Appendix[PDF - 245 KB - 4 pages]). Similarly, LDH and CPK values at 7 dpi correlated significantly with time to death and with log10 viral RNA (Technical Appendix[PDF - 245 KB - 4 pages]).
ROC Curve Analyses for Assessing Clinical Laboratory Values as Early Indicators of EBOV Infection
In the following datasets, ROC curve analysis yielded the best available laboratory predictors as signs of EBOV infection at 3, 5, and 7 dpi: log10 RNA, AST, ALT, CRP, LDH, CPK, and Hgb (Technical Appendix[PDF - 245 KB - 4 pages] Table 2). log10 concentrations of viral RNA outperformed all other individual laboratory values as a predictor of EBOV infection. However, ROC AUC values for log10 RNA were only slightly better than those for LDH, CRP, and AST (Technical Appendix[PDF - 245 KB - 4 pages] Table 2). In fact, when the 3 chemistries were combined (AST+LDH+CRP), they performed almost as well as log10 RNA values in all 3 NHP models. When we compared the combined predictor AST+LDH+CRP−Hgb with log10 RNA values at 3 dpi, it outperformed log10 RNA in all 3 NHP models. For example, at 3 dpi in the EBOV Kikwit–infected rhesus macaque model, ROC AUC was 0.83 for log10 RNA and 0.93 for the combined predictor AST+LDH+CRP−Hgb.
Unlike some other reports showing abnormal laboratory values in NHP EVD models, we have presented our findings in a systematic format concentrating on laboratory values that we think reflect EVD pathogenesis, are easily translatable to human disease, and are potentially available in the human clinical setting. A better understanding of EBOV NHP models will enhance characterization of the disease and facilitate standardization of the models to support possible future vaccine and therapeutic drug submissions under the Food and Drug Administration Animal Rule (https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM399217.pdf). Similar to what has been reported in human EVD (3–12), our findings demonstrate that a lower plasma concentration of viral RNA predicted increased survival time in the 3 NHP models we assessed.
Marzi et al. (23) observed that disease progression in EBOV Makona–infected cynomolgus macaques was delayed compared with that in cynomolgus macaques infected with the EBOV Mayinga strain. Wong et al. (24) compared infections with EBOV Kikwit with infections with 2 different EBOV Makona strains in rhesus macaques and found that the EBOV Makona strains were either similar in virulence or more virulent than the EBOV Kikwit strain they were using. We observed that disease progression in EBOV Makona–infected rhesus macaques was delayed compared with that in EBOV Kikwit–infected rhesus macaques; the observation was supported by viral load data, clinical assessments, and laboratory values. This finding might suggest that, in NHPs, the EBOV Makona strain we used is somewhat less virulent than the EBOV Kikwit strain we used; although, in rhesus macaques, 1,000 PFU of EBOV Makona still resulted in death among all untreated animals. Data are insufficient to determine the relative pathogenicity of EBOV Makona in comparison with that of other EBOV strains. To make this determination, further studies are needed, taking into consideration different EBOV strains and quasispecies.
The difference in survival time between EBOV Kikwit–infected rhesus and cynomolgus macaques (11.7 hours) was not significant; rhesus macaques survived longer. Of interest, at 7 dpi, cynomolgus macaques demonstrated increased impairment of renal function (as determined by BUN and creatinine levels) compared with that for rhesus macaques. This finding does not appear to be associated with a significant observable difference in oral fluid consumption between the animals, and the pathogenesis merits further study.
The search for reliable biomarkers for early diagnosis of EBOV infection and predictors of survival has been a high priority (25,26), but reverse transcription PCR (RT-PCR) remains the reference standard for EVD diagnosis (27). ROC analysis demonstrated that, in all 3 NHP models, qRT-PCR outperformed all individual laboratory values with regard to EVD confirmation. It is noteworthy that the combination of AST+LDH+CRP−Hgb values outperformed qRT-PCR as a laboratory sign of Ebola virus infection at 3 dpi in all 3 NHP models. This could be an important finding and potentially serve as a trigger to treat NHPs in therapeutic studies of IM administered EBOV Kikwit.
Currently, there are no standardized triggers for the initiation of treatment of EBOV-infected NHPs in therapeutics studies. Various time points after virus exposure have been used for therapeutics initiation in NHP models of EBOV infection (28–30). One study used a positive RT-PCR result plus documented fever of >1.5°C above baseline for 1 hour as a prespecified trigger to treat (31). However, it is logistically difficult to obtain timely PCR results in a Biosafety Level 4 laboratory (especially for a large study), and implantation of a telemetry device would be required for optimal fever detection. Thus, standard clinical laboratory values may be a more practical trigger to treat. For example, a calculator (spreadsheet or smartphone application) could be developed to calculate a combined variable value (e.g., AST+LDH+CRP−Hgb) from clinical laboratory values. This approach would be similar to a disease severity smartphone application advocated by Colubri et al. (32) for use with human EBOV patients. Once the threshold laboratory value is reached or exceeded, the therapeutic could be initiated. We intend to conduct a follow-up ROC AUC analysis with a larger sample size to further validate and optimize these preliminary findings.
Our finding that, in lieu of viral load, laboratory values at 5 dpi could potentially predict survival duration is not entirely surprising given that Warren et al. (28) published NHP data that showed the course of EBOV viral load is mirrored by the course of clinical chemistries in the setting of successful EVD treatment using the nucleotide prodrug GS-5734. Although it has been shown that AST levels can predict survival in EBOV-infected humans (5,10,11), we found that LDH may be a better predictor of survival time in NHP models using IM administered EBOV. In all 3 models in our study, LDH and viral load significantly increased at 5 dpi in EBOV Kikwit–infected rhesus and cynomolgus macaques and at 7 dpi in EBOV Makona–infected rhesus macaques. In the EBOV Kikwit–infected rhesus macaque model, LDH values at 5 dpi correlated with viral load and survival time at 5 dpi. Incidentally, LDH has been shown to correlate with survival time in humans with Crimean-Congo hemorrhagic fever, severe fever with thrombocytopenia syndrome, and Dengue virus infection (33–36).
LDH is abundant in the cytoplasm of all human cells and helps catalyze the conversion of pyruvate to lactate, the last step of glycolysis (37). Markedly elevated levels of LDH are often seen in association with cardiogenic shock; hepatic ischemia or necrosis; and intestinal and/or mesenteric ischemia or necrosis (36,38). However, no signs of cardiogenic shock, severe chemical transaminitis corresponding to hepatic ischemia or necrosis, or severe hemolysis were observed at 5 dpi in EBOV Kikwit–infected NHPs or at 7 dpi in EBOV Makona–infected NHPs. One possibility is that the NHPs were experiencing either intestinal and/or mesenteric ischemia or necrosis, conditions that have been seen in other viral infections (39,40) and which may, as postulated by Lynn (41), precipitate late Ebola sepsis-like syndrome.
Kortepeter et al. (42) reported that, in a rhesus macaque model, animals lethally challenged with EBOV Kikwit experienced a rapid increase in plasma viral RNA beginning at 4 dpi and a rapid increase in serum lactate beginning at 7 dpi. In humans, serum lactate levels have been shown to correlate with serum LDH levels (43), and both have independently been associated with death (44–47). Thus, further study is needed of lactate and LDH levels in humans and NHPs with EVD.
A key caveat to our analysis is that our data reflect a retrospective analysis of NHPs used as controls in 5 different studies of IM administered EBOV. The trajectory of early increases in clinical laboratory values, especially CPK and LDH, could be affected by the IM route of EBOV administration. However, Johnson et al. (48) reported similar early increases in LDH and CPK in rhesus macaques in an aerosol model of EBOV Zaire infection. In future analyses, we intend to explore how the route of EBOV administration affects changes in clinical laboratory values. Another limitation to this analysis is that survival time was determined to be time to euthanasia. The strict adherence to the USAMRIID euthanasia criteria, in the setting of 3 clinical assessments daily, supports the supposition that time to euthanasia approximates time to death. Another limitation is that, in this dataset, all animals died, so we were only able to look at survival time. Therefore, we were unable to derive odds ratios for individual variables to assess predictors of death. Future analyses of datasets that include NHPs that survived EBOV infection, with or without treatment with a therapeutic product, will be useful to identify such predictors. Although our analysis included a large number of NHPs (N = 30), we also acknowledge that this retrospective analysis was primarily hypothesis-generating and that there were no prespecified hypotheses. We used p value thresholds of <0.05 for our statistical tests and included an adjusted p value of <0.001 on the basis of a simplified Bonferroni correction for multiple comparisons.
Much can be learned from a critical analysis of EBOV NHP models. Our data support the finding that the virulence of the EBOV Makona strain used in our study may be decreased as compared to that of the EBOV Kikwit strain we used. We did not find a statistically significant difference in survival time when comparing rhesus to cynomolgus macaques in the EBOV Kikwit model, although there were subtle differences in some of the laboratory values. In addition, our data support EBOV studies in humans that indicate basic laboratory values could potentially be used as surrogate markers for viral load and, thus, disease detection and survival. However, validation of this approach in the human clinical setting would also require a comparison with clinical laboratory values associated with endemic diseases present in a given geographic area, such as malaria, rickettsial illnesses, and diseases caused by other hemorrhagic fever viruses. In addition, a combined score of AST, LDH, CRP, and Hgb values could be further evaluated as a trigger to treat NHPs in therapeutics studies of IM administered EBOV. Further work in the NHP model of EVD with regard to clinical and laboratory markers would ideally lead to improvements in predicting survival time in EBOV-infected NHPs and enhancements in the treatment of disease in NHPs, with potential applicability to the management of human EVD.
Dr. Reisler is a lead clinical research contract physician in the Division of Medicine at USAMRIID. His current areas of research interest are biothreat agent research, animal models, vaccine development, and development of therapeutic interventions.
References
- Centers for Disease Control and Prevention. 2014 Ebola outbreak in West Africa—case counts [cited 2016 Dec 5]. https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html
- Kortepeter MG, Bausch DG, Bray M. Basic clinical and laboratory features of filoviral hemorrhagic fever. J Infect Dis. 2011;204(Suppl 3):S810–6. DOIPubMed
- Faye O, Andronico A, Faye O, Salje H, Boëlle PY, Magassouba N, et al. Use of viremia to evaluate the baseline case fatality ratio of Ebola virus disease and inform treatment studies: a retrospective cohort study. PLoS Med. 2015;12:e1001908. DOIPubMed
- Towner JS, Rollin PE, Bausch DG, Sanchez A, Crary SM, Vincent M, et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol. 2004;78:4330–41. DOIPubMed
- Schieffelin JS, Shaffer JG, Goba A, Gbakie M, Gire SK, Colubri A, et al.; KGH Lassa Fever Program; Viral Hemorrhagic Fever Consortium; WHO Clinical Response Team. Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med. 2014;371:2092–100. DOIPubMed
- de La Vega M-A, Caleo G, Audet J, Qiu X, Kozak RA, Brooks JI, et al. Ebola viral load at diagnosis associates with patient outcome and outbreak evolution. J Clin Invest. 2015;125:4421–8. DOIPubMed
- Lanini S, Portella G, Vairo F, Kobinger GP, Pesenti A, Langer M, et al.; INMI-EMERGENCY EBOV Sierra Leone Study Group. Blood kinetics of Ebola virus in survivors and nonsurvivors. J Clin Invest. 2015;125:4692–8. DOIPubMed
- Crowe SJ, Maenner MJ, Kuah S, Erickson BR, Coffee M, Knust B, et al. Prognostic indicators for Ebola patient survival. Emerg Infect Dis. 2016;22:217–23. DOIPubMed
- Fitzpatrick G, Vogt F, Moi Gbabai OB, Decroo T, Keane M, De Clerck H, et al. The contribution of Ebola viral load at admission and other patient characteristics to mortality in a Médecins Sans Frontières Ebola case management centre, Kailahun, Sierra Leone, June–October 2014. J Infect Dis. 2015;212:1752–8. DOIPubMed
- Hunt L, Gupta-Wright A, Simms V, Tamba F, Knott V, Tamba K, et al. Clinical presentation, biochemical, and haematological parameters and their association with outcome in patients with Ebola virus disease: an observational cohort study. Lancet Infect Dis. 2015;15:1292–9. DOIPubMed
- Janvier F, Foissaud V, Cotte J, Aletti M, Savini H, Cordier PY, et al.; Healthcare Workers Ebola Treatment Center Medical Team. Monitoring of prognostic laboratory markers in Ebola virus disease. J Infect Dis. 2016;213:1049. DOIPubMed
- Haaskjold YL, Bolkan HA, Krogh KØ, Jongopi J, Lundeby KM, Mellesmo S, et al. Clinical features of and risk factors for fatal Ebola virus disease, Moyamba District, Sierra Leone, December 2014–February 2015. Emerg Infect Dis. 2016;22:1537–44. DOIPubMed
- Xu Z, Jin B, Teng G, Rong Y, Sun L, Zhang J, et al. Epidemiologic characteristics, clinical manifestations, and risk factors of 139 patients with Ebola virus disease in western Sierra Leone. Am J Infect Control. 2016;44:1285–90. DOIPubMed
- Wong JY, Zhang W, Kargbo D, Haque U, Hu W, Wu P, et al. Assessment of the severity of Ebola virus disease in Sierra Leone in 2014-2015. Epidemiol Infect. 2016;144:1473–81. DOIPubMed
- Qin E, Bi J, Zhao M, Wang Y, Guo T, Yan T, et al. Clinical features of patients with Ebola virus disease in Sierra Leone. Clin Infect Dis. 2015;61:491–5. DOIPubMed
- Sadek RF, Khan AS, Stevens G, Peters CJ, Ksiazek TG. Ebola hemorrhagic fever, Democratic Republic of the Congo, 1995: determinants of survival. J Infect Dis. 1999;179(Suppl 1):S24–7. DOIPubMed
- Agua-Agum J, Ariyarajah A, Blake IM, Cori A, Donnelly CA, Dorigatti I, et al.; WHO Ebola Response Team. Ebola virus disease among male and female persons in West Africa. N Engl J Med. 2016;374:96–8. DOIPubMed
- Rollin PE, Bausch DG, Sanchez A. Blood chemistry measurements and D-Dimer levels associated with fatal and nonfatal outcomes in humans infected with Sudan Ebola virus. J Infect Dis. 2007;196(Suppl 2):S364–71. DOIPubMed
- Barry M, Touré A, Traoré FA, Sako FB, Sylla D, Kpamy DO, et al. Clinical predictors of mortality in patients with Ebola virus disease. Clin Infect Dis. 2015;60:1821–4. DOIPubMed
- Barry M, Traoré FA, Sako FB, Kpamy DO, Bah EI, Poncin M, et al. Ebola outbreak in Conakry, Guinea: epidemiological, clinical, and outcome features.Med Mal Infect. 2014;44:491–4. DOIPubMed
- Martins K, Cooper C, Warren T, Wells J, Bell T, Raymond J, et al. Characterization of clinical and immunological parameters during Ebola virus infection of rhesus macaques. Viral Immunol. 2015;28:32–41. DOIPubMed
- Warren TK, Trefry JC, Marko ST, Chance TB, Wells JB, Pratt WD, et al. Euthanasia assessment in ebola virus infected nonhuman primates. Viruses. 2014;6:4666–82. DOIPubMed
- Marzi A, Feldmann F, Hanley PW, Scott DP, Günther S, Feldmann H. Delayed disease progression in cynomolgus macaques infected with Ebola virus Makona strain. Emerg Infect Dis. 2015;21:1777–83. DOIPubMed
- Wong G, Qiu X, de La Vega MA, Fernando L, Wei H, Bello A, et al. Pathogenicity comparison between the Kikwit and Makona Ebola virus variants in rhesus macaques. J Infect Dis. 2016;214(suppl 3):S281–9. DOIPubMed
- McElroy AK, Spiropoulou CF. Biomarkers for understanding Ebola virus disease. Biomarkers Med. 2014;8:1053–6. DOIPubMed
- Villinger F, Rollin PE, Brar SS, Chikkala NF, Winter J, Sundstrom JB, et al. Markedly elevated levels of interferon (IFN)-γ, IFN-α, interleukin (IL)-2, IL-10, and tumor necrosis factor-α associated with fatal Ebola virus infection. J Infect Dis. 1999;179(Suppl 1):S188–91. DOIPubMed
- Leroy EM, Baize S, Lu CY, McCormick JB, Georges AJ, Georges-Courbot MC, et al. Diagnosis of Ebola haemorrhagic fever by RT-PCR in an epidemic setting. J Med Virol. 2000;60:463–7. DOIPubMed
- Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–5. DOIPubMed
- Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–905. DOIPubMed
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp.Nature. 2014;514:47–53.PubMed
- Pettitt J, Zeitlin L, Kim DH, Working C, Johnson JC, Bohorov O, et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med. 2013;5:199ra113. DOIPubMed
- Colubri A, Silver T, Fradet T, Retzepi K, Fry B, Sabeti P. Transforming clinical data into actionable prognosis models: machine-learning framework and field-deployable app to predict outcome of Ebola patients. PLoS Negl Trop Dis. 2016;10:e0004549. DOIPubMed
- Hasanoglu I, Guner R, Carhan A, Kocak Tufan Z, Yagci-Caglayik D, Guven T, et al. Crucial parameter of the outcome in Crimean Congo hemorrhagic fever: Viral load. J Clin Virol. 2016;75:42–6. DOIPubMed
- Ozturk B, Tutuncu E, Kuscu F, Gurbuz Y, Sencan I, Tuzun H. Evaluation of factors predictive of the prognosis in Crimean-Congo hemorrhagic fever: new suggestions. Int J Infect Dis. 2012;16:e89–93. DOIPubMed
- Gai ZT, Zhang Y, Liang MF, Jin C, Zhang S, Zhu CB, et al. Clinical progress and risk factors for death in severe fever with thrombocytopenia syndrome patients. J Infect Dis. 2012;206:1095–102. DOIPubMed
- Sirikutt P, Kalayanarooj S. Serum lactate and lactate dehydrogenase as parameters for the prediction of dengue severity. J Med Assoc Thai. 2014;97(Suppl 6):S220–31.PubMed
- Adeva-Andany M, López-Ojén M, Funcasta-Calderón R, Ameneiros-Rodríguez E, Donapetry-García C, Vila-Altesor M, et al. Comprehensive review on lactate metabolism in human health. Mitochondrion. 2014;17:76–100. DOIPubMed
- Guzmán-de la Garza FJ, Ibarra-Hernández JM, Cordero-Pérez P, Villegas-Quintero P, Villarreal-Ovalle CI, Torres-González L, et al. Temporal relationship of serum markers and tissue damage during acute intestinal ischemia/reperfusion. Clinics (Sao Paulo). 2013;68:1034–8. DOIPubMed
- Yasuda H, Okita Y, Imaoka H, Fujikawa H, Ohi M, Araki T, et al. Intestinal necrosis due to norovirus enteritis. Clin J Gastroenterol. 2015;8:10–3. DOIPubMed
- de Villiers FPR, Driessen M. Clinical neonatal rotavirus infection: association with necrotising enterocolitis. S Afr Med J. 2012;102:620–4. DOIPubMed
- Lynn LA. Combined endothelial and epithelial barrier disruption of the colon may be a contributing factor to the Ebola sepsis-like syndrome. Patient Saf Surg. 2015;9:1. DOIPubMed
- Kortepeter MG, Lawler JV, Honko A, Bray M, Johnson JC, Purcell BK, et al. Real-time monitoring of cardiovascular function in rhesus macaques infected with Zaire ebolavirus. J Infect Dis. 2011;204(Suppl 3):S1000–10. DOIPubMed
- Zein JG, Lee GL, Tawk M, Dabaja M, Kinasewitz GT. Prognostic significance of elevated serum lactate dehydrogenase (LDH) in patients with severe sepsis. Chest. 2004;126(4_Meeting Abstracts):873S. DOI
- Kruse O, Grunnet N, Barfod C. Blood lactate as a predictor for in-hospital mortality in patients admitted acutely to hospital: a systematic review.Scand J Trauma Resusc Emerg Med. 2011;19:74. DOIPubMed
- Vincent JL, Quintairos E Silva A, Couto L Jr, Taccone FS. The value of blood lactate kinetics in critically ill patients: a systematic review. Crit Care. 2016;20:257. DOIPubMed
- Duman A, Akoz A, Kapci M, Ture M, Orun S, Karaman K, et al. Prognostic value of neglected biomarker in sepsis patients with the old and new criteria: predictive role of lactate dehydrogenase. Am J Emerg Med. 2016;34:2167–71. DOIPubMed
- Erez A, Shental O, Tchebiner JZ, Laufer-Perl M, Wasserman A, Sella T, et al. Diagnostic and prognostic value of very high serum lactate dehydrogenase in admitted medical patients. Isr Med Assoc J. 2014;16:439–43.PubMed
- Johnson E, Jaax N, White J, Jahrling P. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int J Exp Pathol. 1995;76:227–36.PubMed





















%205.10.48%E2%80%AFa.m..png)












No hay comentarios:
Publicar un comentario