
Volume 26, Number 6—June 2020
Research
Radical Change in Zoonotic Abilities of Atypical BSE Prion Strains as Evidenced by Crossing of Sheep Species Barrier in Transgenic Mice
Article Metrics
Alba Marín-Moreno1, Alvina Huor1, Juan Carlos Espinosa, Jean Yves Douet, Patricia Aguilar-Calvo2, Naima Aron, Juan Píquer, Sévérine Lugan, Patricia Lorenzo, Cecile Tillier, Hervé Cassard, Olivier Andreoletti, and Juan María Torres
Abstract
Classical bovine spongiform encephalopathy (BSE) is the only zoonotic prion disease described to date. Although the zoonotic potential of atypical BSE prions have been partially studied, an extensive analysis is still needed. We conducted a systematic study by inoculating atypical BSE isolates from different countries in Europe into transgenic mice overexpressing human prion protein (PrP): TgMet129, TgMet/Val129, and TgVal129. L-type BSE showed a higher zoonotic potential in TgMet129 mice than classical BSE, whereas Val129-PrP variant was a strong molecular protector against L-type BSE prions, even in heterozygosis. H-type BSE could not be transmitted to any of the mice. We also adapted 1 H- and 1 L-type BSE isolate to sheep-PrP transgenic mice and inoculated them into human-PrP transgenic mice. Atypical BSE prions showed a modification in their zoonotic ability after adaptation to sheep-PrP producing agents able to infect TgMet129 and TgVal129, bearing features that make them indistinguishable of sporadic Creutzfeldt-Jakob disease prions.
Prion diseases, or transmissible spongiform encephalopathies (TSEs), are a group of rare and lethal neurodegenerative diseases that affect a great number of mammal species, including humans and animals belonging to the human food chain. The conversion of a host encoded cellular protein of unknown function (PrPC) into a disease-associated isoform (PrPSc) is the molecular event underlying the development of TSEs. Such conformational change is driven by PrPSc itself because it recruits and transforms PrPC molecules, acting as a template (1). The conformational change, associated with an increase of β-sheet content, also results in a change in the protein biochemical features (2). Although PrPC is monomeric, protease-sensitive, and soluble in nonionic detergents, PrPSc has a high tendency to aggregate, is partially resistant to protease digestion, and is insoluble in nonionic detergents (1,3).
Classical bovine spongiform encephalopathy (C-BSE) caused a major food safety crisis when consumption of contaminated meat was discovered in the late 1990s as the cause of a new prion disease affecting humans, which was called variant Creutzfeldt-Jakob disease (vCJD) (4). The first description of C-BSE was made in 1987 in affected cattle in the United Kingdom (5). In the years following, ≈200,000 cattle succumbed to the disease (6). To date, C-BSE is the only recognized zoonotic prion (6) and is responsible for >231 human deaths (7).
After the implementation of active surveillance in the European Union in 2001, several atypical BSE cases were identified in aged asymptomatic cattle during slaughterhouse testing. Two major phenotypes with pathology and epidemiology distinct from C-BSE were observed, bovine amyloidotic spongiform encephalopathy (or L-BSE) (8) and H-BSE (9). The biochemical properties of PrPSc isolated from these cases also differed from C-BSE in terms of the protease-resistant fragment size and ratio of glycoforms on Western blot (WB). It is unclear whether atypical BSE resulted from exposure to an acquired TSE or emerged spontaneously, a theory supported by the occurrence of atypical BSE being maintained at a similar rate in various countries independent of their C-BSE status.
The zoonotic potentials of atypical and C-BSE seemed to differ. One study performed in transgenic mice overexpressing the human Met129–normal brain prion protein (PrP) variant (Tg650) showed that L-BSE has a higher zoonotic potential than C-BSE because a 100% attack rate was observed in the intracranial challenge, whereas H-BSE was unsuccessfully transmitted (10). Other intermediate species belonging to the human food chain might play a role in a possible atypical BSE zoonosis. For example, C-BSE can naturally affect goats (11). Furthermore, C-BSE virulence in human-PrP transgenic mouse models is increased after passaging in ovine-PrP transgenic mice (12). The possible zoonotic potential of atypical BSE after its adaptation to the sheep sequence is not known. At least L-type BSE is efficiently transmitted in sheep (13). L-BSE transmission in ovine-PrP transgenic mouse models produced a prion similar to C-BSE in terms of incubation times, histopathologic features, and electrophoretic mobility, although the glycoprofile maintained a more equilibrated proportion between the 3 bands than C-BSE (14). Therefore, a deep assessment of the zoonotic potential of atypical BSE prions should include the evaluation of zoonotic potential after adaptation to the sheep-PrP sequence.
Polymorphisms and mutations in the human prion protein gene affect survival and disease development in vCJD and other human TSEs (15). The most important genetic variant for disease outcome in humans is the polymorphic codon 129, which can codify methionine (Met129) or valine (Val129) and has been detected as Met129 homozygous in all vCJD-diagnosed cases, with the exception of 1 Met/Val129 heterozygous vCJD case (16,17).
The main aim of our study was to assess the zoonotic potential of the atypical BSE prions in transgenic mice that overexpress human-PrP covering the 3 possible 129 codon genotypes. We used a high number of isolates from different sites in Europe and a collection of human-PrP transgenic mouse lines. In addition, we adapted 1 H-BSE and 1 L-BSE isolate to the sheep-PrP sequence and sequentially inoculated them into human-PrP TgMet129 and TgVal129 mice to assess whether intermediate passage in sheep can modify prion strain features, including prion virulence in humans. We decided to use 2 different ovine-PrP transgenic mouse models to study the effect of the polymorphism Ala/Val136 of the sheep-PrP sequence.
Ethics Statement
We conducted animal experiments in accordance with the Code for Methods and Welfare Considerations in Behavioral Research with Animals (Directive 2010/63/EU) and made every effort to minimize suffering. Experiments developed in Centro de Investigación en Sanidad Animal–Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (Madrid, Spain) were evaluated by the Committee on the Ethics of Animal Experiments of the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria and approved by the General Directorate of the Madrid Community Government (permit nos. PROEX 263/15, PROEX 181/16, and PROEX 228/16). Experiments developed in Institut National de la Recherche Agronomique–École Nationale Vétérinaire de Toulouse (Toulouse, France) were approved by the Institut National de la Recherche Agronomique/École Nationale Vétérinaire de Toulouse Ethics Committee under the auspices of the Ministry of Education and Research of France (permit no. APAFIS-2017044210492380 v2).
Prion Isolates
We used a collection of atypical BSE field isolates from different countries in Europe to ensure the consistency of the results (Appendix Table). We characterized all isolates to differentiate them from C-BSE (data not shown). For comparison, we included C-BSE isolates and other prion isolates in the study. For inoculation, we prepared all isolates from brain tissues as 10% (wt/vol) homogenates in 5% glucose. Second passages were performed by inoculating mice with 10% (wt/vol) homogenates in 5% glucose of brains selected from first passage.
Mouse Transmission Studies
The atypical BSE isolates were inoculated in 3 different mouse models for human-PrP: HuPrP-Tg340-Met129 (TgMet129) line expressing human Met129-PrP variant (12), HuPrP-Tg361-Val129 (TgVal129) line expressing human Val129-PrP variant (18), and HuPrP-Tg351-Met/Val129 (TgMet/Val129) line obtained by mating TgMet129 and TgVal129 mice (18). All of these transgenic lines show similar brain PrPC levels of expression (≈4-fold the level of expression in human brain) on a mouse-PrP null background. For the adaptation to the sheep-PrP sequence, we used 2 different overexpressing sheep-PrP mouse models to include the 2 variants of the Ala/Val136 Arg154 Gln171 haplotype. The OvPrP-Tg338 (TgVRQ) harbors the VRQ allele and expresses 8–10-fold the level of PrPC expression in sheep brain (19). The OvPrP-TgShXI (TgARQ) harbors the ARQ allele and expresses 3–4-fold the level of PrPC expression in sheep brain (20). We performed subsequent bioassays for the detection of low-level propagation of atypical BSE prions in BoPrP-Tg110 mice (TgBo) (21).
We individually anesthetized 6–7-week-old mice with isoflurane and inoculated them with 2 mg equivalent of brain homogenate in the right parietal lobe by using a 25-gauge disposable hypodermic needle. We observed mice daily and assessed their neurologic status twice a week. When progression of a TSE disease was evident or at the established experimental endpoint (700 days postinoculation), we euthanized the animals for ethical reasons, then performed necropsy and removed the brain. We fixed part of the brain by using immersion in neutral-buffered 10% formalin (4% 2-formaldehyde) and used it for histopathology; we froze the rest of the tissue at −20°C and used it for determining the presence of proteinase K–resistant PrPSc (PrPres) by WB. We calculated survival times as mean + SD days postinoculation for all the mice that scored positive for PrPres. We defined the attack rate as the proportion of mice that scored positive for PrPres divided by the number of inoculated mice. We used brain homogenates from PrPres-positive mice for further passaging. When all mice were scored negative for PrPres on primary passage, we pooled all PrPres-negative brains and used them for second passage.
Histologic Examination and Paraffin-Embedded Tissue Blotting
We performed all procedures comprising the histopathologic analysis of mouse brains as previously described (22). We fixed mouse brain samples in neutral-buffered 10% formalin (4% 2-formaldehyde) and embedded them in paraffin. After deparaffinization, we stained 4-µm-thick tissue slices with hematoxylin and eosin. We established brain lesion profiles according to published methods (23). We conducted paraffin-embedded tissue (PET) blots as previously described (24) by using the Sha31 monoclonal antibody (mAb) (25).
Western Blotting
We homogenized frozen brain tissues (175 + 20 mg) in 5% glucose in distilled water in grinding tubes (Bio-Rad, ) adjusted to 10% (wt/vol) by using a TeSeE Precess 48TM homogenizer (Bio-Rad). We determined the presence of PrPres in transgenic mouse brains by using WB, using the reagents of the ELISA commercial test TeSeE (Bio-Rad). We prepared brain homogenates (10–100 µL of a 10% [wt/vol]) as previously described (18) and loaded samples into 12% Bis-Tris Gel (Criterion XT; Bio-Rad). We transferred proteins electrophoretically onto polyvinylidene fluoride membranes (Millipore, ) and blocked overnight with 2% bovine serum albumin blocking buffer. We incubated membranes with Sha31 (25) mAb at a concentration of 1 µg/mL. Sha31 recognizes the 145-WEDRYYRE-152 epitope of the human-PrPC sequence, which is conserved in sheep and bovine sequences. We detected immunocomplexes by incubating the membranes for 1 h with horseradish peroxidase conjugated antimouse IgG (GE Healthcare Amersham Biosciences, ). We developed immunoblots with enhanced chemiluminescence in ECL Select (GE Healthcare Amersham Biosciences) and captured images by using the ChemiDoc WRS+ System and processed them by using Image Lab 5.2.1 software (both Bio-Rad).
Atypical BSE Transmission to Human-PrP Transgenic Mouse Models
We transmitted a panel of atypical L-type and H-type BSE (2 serial passages) into 3 transgenic mouse lines. These mouse lines were homozygous for Met (TgMet129) or Val (TgVal129) at codon 129 of human-PrP or were their F1 cross resulting in heterozygous mice (TgMet/Val129). The PrPC level in the brain of all 3 transgenic mice lines has been shown to be approximately 4-fold higher than in human brain tissue (26). In parallel, we inoculated C-BSE control isolates (Table 1).
As we reported on a previous study (18), only TgMet129 mice get infected with C-BSE isolates. However, TgMet/Val129 and TgVal129 showed no transmission or accumulation of PrPres (Table 1).
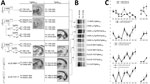
L-BSE was efficiently transmitted to TgMet129 (Table 1; Figure 1, panel A). The attack rate was 100% for first passage, and incubation time was not reduced on subsequent passaging. In TgMet129, the L-BSE PrPres pattern in WB differed from the C-BSE PrPres pattern both in terms of apparent molecular weight and glycoprofile distribution, marked by an evident lower proportion of the diglycosylated band (Figure 1, panel B; Appendix Figure 1). We examined the regional distribution and intensity of PrPres deposition in the brain by using PET blotting. Brains of TgMet129 inoculated with L-BSE showed finer staining than the granular PrP deposits typical of C-BSE (Figure 1; Appendix Figure 1). Deposits were mostly restricted to the habenular, geniculate, and dorsal nuclei of the thalamus. Lesion profiles also reflect differences between both strains (Figure 1, panel C). We detected no clinical signs, PrPres accumulation by WB (Table 1), or PrPSc deposition by PET blotting (data not shown) in the brains of TgVal129 or TgMet/Val129 L-BSE inoculated mice. These brains were collected and inoculated in TgBo animals; no transmission was observed (data not shown). This finding suggests the absence of subclinical infection in TgVal129 and TgMet/Val129 mice. We inoculated L-BSE passaged in TgMet129 into TgVal129 and TgMet/Val129 mice and detected no PrPres accumulation (Table 1). After H-BSE inoculation, we detected no clinical signs, PrPres accumulation by WB (Table 1), or PrPSc deposits by PET blotting (data not shown) in any of the 3 human transgenic mouse models.
Atypical BSE Transmission into Ovine-PrP Transgenic Mouse Models
We transmitted 1 L-BSE isolate and 1 H-BSE isolate (2 iterative passages) to VRQ and ARQ ovine-PrP transgenic mice. L-BSE transmitted in both TgVRQ and TgARQ caused 100% attack rates and short incubation times in the second passage (Table 2).
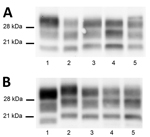
L-BSE propagation in all TgVRQ and TgARQ infected mice showed a WB PrPres profile with similarities to the one observed after passage of C-BSE in the same model in terms of low molecular mass migration but also a glycoform ratio where the diglycosylated and monoglycosylated bands contain a more equal signal proportion than C-BSE were detected with mAb Sha31, which is more similar to L-BSE or classical scrapie (Figure 2, panel A). These features have previously been reported in ARQ/ARQ and VRQ/VRQ sheep inoculated with L-BSE (13). We inoculated brains collected from the second passage in TgBo, causing a clinical disease with similar incubation periods as original L-BSE (Table 3). Both WB and PET blotting patterns (Appendix Figure 2), as well as WB patterns once transmitted back into TgBo (Figure 3), support the view that passage in ovine sequences did not irreversibly alter L-BSE strain properties.
TgARQ mice inoculated with H-BSE isolate did not show any clinical sign or accumulation of PrPres (Table 2). In TgVRQ, first passage produced disease in only 1 out of 6 animals, with long incubation times. On second passage, 100% attack rates were achieved, and incubation times were reduced (Table 2). This finding suggests a substantial transmission barrier for the H-BSE prion agent in this animal model. The WB PrPres profile was characterized by 21 kDa profile (Figure 2, panel B). The prion that propagated in TgVRQ mice inoculated with H-BSE could not be transmitted (2 serial passages) in TgBo mice. These results suggest that, upon adaptation to the VRQ ovine-PrP sequence, the H-BSE strain properties were irreversibly altered (Table 3).
We thus analyzed the neuropathologic phenotypes of the atypical BSE agents transmitted into ovine-PrP transgenic mice by PrPres PET blotting (Appendix Figure 2). Regarding L-BSE, the results support those obtained by WB because TgVRQ ovine passaged L-BSE displays a similar PrP deposition as ovine passaged C-BSE. PrPres deposition predominantly involved several nuclei of the thalamus and regions like the septum and external cortex of the inferior colliculus; we obtained similar results for TgARQ passaged L-BSE (Appendix Figure 2), as previously described (14). Concerning TgVRQ adapted H-BSE prions, the deposition pattern is also strikingly different to that displayed by C-BSE or L-BSE (Appendix Figure 2).
Atypical BSE Transmission into Human-PrP Transgenic Mouse Models after Adaptation to the Sheep-PrP Sequence
We inoculated prions obtained after 2 passages of L-BSE and H-BSE isolates in ovine PrP expressing mice into TgMet129 and TgVal129. TgMet129 mice inoculated with L-BSE passaged into TgVRQ showed neither clinical signs nor PrPres accumulation in their brain (Figure 1, panels A, B). By contrast, TgVal129 inoculated with the same isolates had disease characterized by 100% attack rates and short incubation times in the second passage (Figure 1, panel A). The obtained PrPres resembles a sporadic Creutzfeldt-Jakob disease (sCJD) Val/Val129 type 2 (sCJD VV2) profile (Figure 1, panel B). PET blotting showed a deposition pattern similar to that of sCJD VV2 in the same mice (Figure 1, panel A). Lesion profiles of both strains also were coincident (Figure 1, panel C). Furthermore, the prion generated in the TgVal129 transgenic mouse line is not able to infect back in TgBo (Table 3). By contrast, ARQ-adapted L-BSE loses its ability to infect human-PrP TgMet129 transgenic mouse lines (Figure 1, panel A).
VRQ-adapted H-BSE can now infect both human-PrP TgMet129 and TgVal129 mouse lines, showing 100% attack rates and short incubation times in the second passage (Figure 1, panel A). The obtained PrPres in both cases is similar to that of sCJD Met/Met129 type 1 (sCJD MM1) (Figure 1, panel B). PET blotting showed similarities with the deposition patterns typical of sCJD MM1 in TgMet129 and TgVal129 (Figure 1, panel A). Lesion profiles of both strains also were coincident (Figure 1, panel C). Both generated prions can infect back in TgBo (Table 3; Figure 3).
The zoonotic potential of atypical BSE prions has been partially studied by using both PrP-overexpressing animals and gene-targeted mice (10,27–29). All evidence converges to indicate a higher capacity of L-BSE than of C-BSE to transmit in human-PrP–expressing hosts and a high transmission barrier for H-BSE. Absence of a transmission barrier for L-BSE in TgMet129 has already been reported (10). Our study (using other transgenic mice with a different PrPC expression level) expands the investigation to other genotypes. Lack of prion propagation in TgMet/Val129 and TgVal129 indicates that Val129 variant is a strong molecular protector against L-BSE zoonotic transmission even in heterozygosis, as previously reported for C-BSE and related C-BSE prions (18). Finally, H-BSE clearly has a robust transmission barrier with respect to the human-PrP sequence, independent of the codon 129 polymorphism.
Once adapted to TgMet129, L-BSE did not propagate in TgMet/Val129 and TgVal129, which precludes secondary infections between persons. In contrast, C-BSE propagated in Val129 genotypes once adapted to the Met129 human-PrP sequence, even when Val129 also protected against primary infection (18). However, Met/Val129 genotypes might be naturally affected by vCJD because a definite vCJD case of a subject heterozygous for codon 129 has already been reported (16). Moreover, evidence of potential prion propagation in Val129-homozygous persons has been indicated in examinations of tonsils and appendices (30–32). These observations are in contrast with our finding of a lack of transmission of C-BSE in Val129 genotypes. However, only 1 vCJD case has been reported in a Met/Val-heterozygous person, which might mean that the event is very rare. Moreover, whether the PrPres positivity detected in lymphoid tissues (tonsils and appendix) of Val129-homozygous persons would eventually extend to their central nervous system is still unknown. As a consequence, the risk for L-BSE secondary transmission once adapted to human-PrP sequence should be assessed carefully.
A complete assessment of the zoonotic potential of atypical BSE prions should include the evaluation of zoonotic potential after adaptation to the sheep-PrP sequence given that C-BSE virulence toward human-PrP transgenic mouse models increased after passage in ovine-PrP transgenic mice. Thus, we propagated 1 H-BSE and 1 L-BSE isolate into ovine-PrP transgenic mice. L-BSE has already been transmitted into sheep (13), whereas no H-BSE propagation into this host has been reported. Thus, we decided to perform the adaptation to the sheep sequence by using sheep-PrP transgenic mice, although overexpression of PrPC reportedly could render changes on prion strains features (33). The absence of divergent PrPres profiles among the animals and the uniformity of the incubation times after 2 passages into sheep-PrP transgenic mice argue against the occurrence of mutation events attributable to PrPC overexpression in our study. In addition, previously reported strain features of L-BSE propagated in sheep (13) were similar to those reported in this study using sheep-PrP transgenic mice, which also validates use of these animal models. Our results suggest a moderate but surmountable transmission barrier for L-BSE in the 2 analyzed sheep genotypes, whereas for H-BSE a high transmission barrier exists when transmitted into an ARQ sheep sequence. The polymorphism Ala/Val136 present in the sheep-PrP sequence seems to be responsible for the different behavior of the obtained prion agents. Once proved that these isolates could be propagated on sheep-PrP sequence, determining whether they can be differentiated from classical scrapie and C-BSE will be important.
Our results and those provided by other studies indicate that L-BSE adapted to a VRQ sheep sequence resemble C-BSE in its molecular features (14). Moreover, L-BSE adapted to ARQ sheep sequence and H-BSE adapted to VRQ sheep sequence generate prions similar to classical scrapie, at least in terms of PrPres glycoprofile. Therefore, in the supposed case of atypical BSE transmission to sheep, early differentiation of both atypical BSE agents from other sheep prions like classical scrapie would be difficult. Nevertheless, the combination of the low incidence of atypical BSE (because of its supposed sporadic nature) and the continued prohibition of meat and bone meal recycling ameliorates the risk for transmission to sheep.
The transmission of atypical BSEs into sheep resulted in the emergence of prions similar to types 1 and 2 sCJD in terms of mean survival times, attack rates, PrPres profile, and PrPres deposition pattern in the brain of human-PrP transgenic mice. The similarities between the sheep-adapted atypical BSE prions propagated into our human-PrP transgenic mouse lines and sCJD prions could suggest a link between them. The well-established dogma that sCJD is a spontaneous disorder unrelated to animal prion disease has been questioned in a previous study given the resemblance of scrapie prions transmitted into human transgenic mouse models to sCJD strains (26); however, the data from that study do not unequivocally establish a causative link between exposure to sheep scrapie and the subsequent appearance of sCJD in humans, and the same could apply to our findings. An alternative explanation that cannot be ruled out is that, although being different strains, only a limited number of phenotypes could be generated for the human-PrP, indicating phenotypic convergence. Updates to old epidemiologic research is needed to reconsider all these results involving a possible infectious origin of sCJD. In any case, continuing the characterization of this newly emerged prion strain would be useful to finally discarding or refuting a link with sCJD prions.
Extrapolation of results from prion transmission studies based on transgenic mice should be done with caution, especially when human susceptibility to prions is analyzed. However, our results clearly indicate that atypical BSE adaptation to an ovine-PrP sequence could modify the prion agent to potentially infect humans, showing strain features indistinguishable from those of classic sCJD prions, even though they might or might not be different agents. The supposed sporadic nature of atypical BSE makes its transmission to sheep and later to humans unlikely. However, the expanding range of TSE agents displaying the capacity to transmit in human-PrP–expressing hosts warrants the continuation of the ban on meat and bone meal recycling and underscores the ongoing need for active surveillance.
Ms. Marín-Moreno is a PhD student in the Prion Group at Centro de Investigación en Sanidad Animal–Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Madrid, Spain. Her primary research interests are the characterization of prion strains and the pathogenesis of prion diseases and their effects on animal and human health.
Acknowledgments
We thank the staff of the Biosafety Level 3 animal facility and the Biosafety Office at Centro de Investigación en Sanidad Animal–Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (Valdeolmos-Madrid) for their excellent animal care and work, and C. Lacroux for her assistance in organizing the mouse bioassays. In addition, we thank the suppliers of the isolates used in this study.
This work was funded by the Food Standards Agency (grant no. FS231051 to O.A. and J.M.T.), Spanish Ministerio de Economía Industria y Competitividad (grant no. RTA2012-00004 to J.M.T, grant no. AGL2016-78054-R [AEI/FEDER, UE] to J.M.T. and J.C.E., and fellowship BES-2010- 040922 to P.A.C.), Fundació La Marató de TV3 (grant no. 201821-31 to J.C.E.), Instituto Nacional de Investigación y Tecnología Agraria y Agroalimentaria (fellowship INIA-FPI-SGIT-2015-02 to A.M.M.), Alliance BioSecure Research Foundation (grant no. FABS FRM-2014 to J.M.T.), and the FEDER programs POCTEFA (EFA148/16 to O.A.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conception and design of the study, as well as analysis and interpretation of data and acquisition of funding, were done by J.M.T, J.C.E., and O.A. All authors contributed to data acquisition. A.M.M, J.M.T, J.C.E., and O.A. were involved in drafting the manuscript and revising it critically for important intellectual content. All authors gave final approval of the version to be published.
References
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83.
- Vázquez-Fernández E, Vos MR, Afanasyev P, Cebey L, Sevillano AM, Vidal E, et al. The structural architecture of an infectious mammalian prion using electron cryomicroscopy. PLoS Pathog. 2016;12:
e1005835 . - Gielbert A, Davis LA, Sayers AR, Hope J, Gill AC, Sauer MJ. High-resolution differentiation of transmissible spongiform encephalopathy strains by quantitative N-terminal amino acid profiling (N-TAAP) of PK-digested abnormal prion protein. J Mass Spectrom. 2009;44:384–96.
- Will RG, Ironside JW, Zeidler M, Cousens SN, Estibeiro K, Alperovitch A, et al. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347:921–5.
- Wells GA, Scott AC, Johnson CT, Gunning RF, Hancock RD, Jeffrey M, et al. A novel progressive spongiform encephalopathy in cattle. Vet Rec. 1987;121:419–20.
- Fernández-Borges N, Marín-Moreno A, Konold T, Espinosa JC, Torres JM. Bovine spongiform encephalopathy (BSE). In: Reference module in neuroscience and biobehavioral psychology. London: Elsevier; 2017. p.1–10.
- Houston F, Andréoletti O. Animal prion diseases: the risks to human health. Brain Pathol. 2019;29:248–62.
- Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, et al. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A. 2004;101:3065–70.
- Biacabe AG, Laplanche JL, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004;5:110–5.
- Béringue V, Herzog L, Reine F, Le Dur A, Casalone C, Vilotte JL, et al. Transmission of atypical bovine prions to mice transgenic for human prion protein. Emerg Infect Dis. 2008;14:1898–901.
- Eloit M, Adjou K, Coulpier M, Fontaine JJ, Hamel R, Lilin T, et al. BSE agent signatures in a goat. Vet Rec. 2005;156:523–4.
- Padilla D, Béringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, et al. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 2011;7:
e1001319 . - Simmons MM, Chaplin MJ, Konold T, Casalone C, Beck KE, Thorne L, et al. L-BSE experimentally transmitted to sheep presents as a unique disease phenotype. Vet Res (Faisalabad). 2016;47:112.
- Béringue V, Andréoletti O, Le Dur A, Essalmani R, Vilotte JL, Lacroux C, et al. A bovine prion acquires an epidemic bovine spongiform encephalopathy strain-like phenotype on interspecies transmission. J Neurosci. 2007;27:6965–71.
- Lloyd SE, Mead S, Collinge J. Genetics of prion diseases. Curr Opin Genet Dev. 2013;23:345–51.
- Mok T, Jaunmuktane Z, Joiner S, Campbell T, Morgan C, Wakerley B, et al. Variant Creutzfeldt-Jakob disease in a patient with heterozygosity at PRNP codon 129. N Engl J Med. 2017;376:292–4.
- Will RG, Zeidler M, Stewart GE, Macleod MA, Ironside JW, Cousens SN, et al. Diagnosis of new variant Creutzfeldt-Jakob disease. Ann Neurol. 2000;47:575–82.
- Fernández-Borges N, Espinosa JC, Marín-Moreno A, Aguilar-Calvo P, Asante EA, Kitamoto T, et al. Val129-PrP variant is a strong molecular protector against BSE zoonotic transmission but fails to prevent human-to-human vCJD transmission. Emerg Infect Dis. 2017;23:1522–30.
- Le Dur A, Béringue V, Andréoletti O, Reine F, Laï TL, Baron T, et al. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc Natl Acad Sci U S A. 2005;102:16031–6.
- Kupfer L, Eiden M, Buschmann A, Groschup MH. Amino acid sequence and prion strain specific effects on the in vitro and in vivo convertibility of ovine/murine and bovine/murine prion protein chimeras. Biochim Biophys Acta. 2007;1772:704–13.
- Castilla J, Gutiérrez Adán A, Brun A, Pintado B, Ramírez MA, Parra B, et al. Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch Virol. 2003;148:677–91.
- Andréoletti O, Berthon P, Levavasseur E, Marc D, Lantier F, Monks E, et al. Phenotyping of protein-prion (PrPsc)-accumulating cells in lymphoid and neural tissues of naturally scrapie-affected sheep by double-labeling immunohistochemistry. J Histochem Cytochem. 2002;50:1357–70.
- Fraser H, Dickinson AG. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol. 1968;78:301–11.
- Andréoletti O, Simon S, Lacroux C, Morel N, Tabouret G, Chabert A, et al. PrPSc accumulation in myocytes from sheep incubating natural scrapie. Nat Med. 2004;10:591–3.
- Féraudet C, Morel N, Simon S, Volland H, Frobert Y, Créminon C, et al. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem. 2005;280:11247–58.
- Cassard H, Torres JM, Lacroux C, Douet JY, Benestad SL, Lantier F, et al. Evidence for zoonotic potential of ovine scrapie prions. Nat Commun. 2014;5:5821.
- Kong Q, Zheng M, Casalone C, Qing L, Huang S, Chakraborty B, et al. Evaluation of the human transmission risk of an atypical bovine spongiform encephalopathy prion strain. J Virol. 2008;82:3697–701.
- Wilson R, Dobie K, Hunter N, Casalone C, Baron T, Barron RM. Presence of subclinical infection in gene-targeted human prion protein transgenic mice exposed to atypical bovine spongiform encephalopathy. J Gen Virol. 2013;94:2819–27.
- Wilson R, Plinston C, Hunter N, Casalone C, Corona C, Tagliavini F, et al. Chronic wasting disease and atypical forms of bovine spongiform encephalopathy and scrapie are not transmissible to mice expressing wild-type levels of human prion protein. J Gen Virol. 2012;93:1624–9.
- Gill ON, Spencer Y, Richard-Loendt A, Kelly C, Dabaghian R, Boyes L, et al. Prevalent abnormal prion protein in human appendixes after bovine spongiform encephalopathy epizootic: large scale survey. BMJ. 2013;347(oct15 5):f5675.
- Hilton DA, Ghani AC, Conyers L, Edwards P, McCardle L, Ritchie D, et al. Prevalence of lymphoreticular prion protein accumulation in UK tissue samples. J Pathol. 2004;203:733–9.
- Ironside JW, Bishop MT, Connolly K, Hegazy D, Lowrie S, Le Grice M, et al. Variant Creutzfeldt-Jakob disease: prion protein genotype analysis of positive appendix tissue samples from a retrospective prevalence study. BMJ. 2006;332:1186–8.
- Le Dur A, Laï TL, Stinnakre MG, Laisné A, Chenais N, Rakotobe S, et al. Divergent prion strain evolution driven by PrPC expression level in transgenic mice. Nat Commun. 2017;8:14170.
Figures
Tables
Cite This ArticleOriginal Publication Date: April 21, 2020
1These first authors contributed equally to this article.
2Current affiliation: University of California–San Diego, La Jolla, California, USA.





















.png)












No hay comentarios:
Publicar un comentario