
Volume 26, Number 3—March 2020
Research
Acquisition of Plasmid with Carbapenem-Resistance Gene blaKPC2 in Hypervirulent Klebsiella pneumoniae, Singapore
Article Metrics
Yahua Chen, Kalisvar Marimuthu, Jeanette Teo, Indumathi Venkatachalam, Benjamin Pei Zhi Cherng, Liang De Wang, Sai Rama Sridatta Prakki, Weizhen Xu, Yi Han Tan, Lan Chi Nguyen, Tse Hsien Koh, Oon Tek Ng, and Yunn-Hwen Gan
Abstract
The convergence of carbapenem-resistance and hypervirulence genes in Klebsiella pneumoniae has led to the emergence of highly drug-resistant superbugs capable of causing invasive disease. We analyzed 556 carbapenem-resistant K. pneumoniae isolates from patients in Singapore hospitals during 2010–2015 and discovered 18 isolates from 7 patients also harbored hypervirulence features. All isolates contained a closely related plasmid (pKPC2) harboring blaKPC-2, a K. pneumoniae carbapenemase gene, and had a hypervirulent background of capsular serotypes K1, K2, and K20. In total, 5 of 7 first patient isolates were hypermucoviscous, and 6 were virulent in mice. The pKPC2 was highly transmissible and remarkably stable, maintained in bacteria within a patient with few changes for months in the absence of antimicrobial drug selection pressure. Intrapatient isolates were also able to acquire additional antimicrobial drug resistance genes when inside human bodies. Our results highlight the potential spread of carbapenem-resistant hypervirulent K. pneumoniae in Singapore.
The rise of multidrug-resistant (MDR) Enterobacteriaceae prompted the World Health Organization to classify carbapenem-resistant Enterobacteriaceae, of which Klebsiella is the most common genus, on the global priority list of antibiotic-resistant bacteria in 2017 (1). Carbapenem-resistant K. pneumoniae (CRKP, also including K. quasipneumoniae) infections are generally hospital acquired, particularly among elderly and immunocompromised patients (2,3). The major carbapenemases include K. pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase, and carbapenem-hydrolyzing class D β-lactamase (OXA), all of which have spread globally (4–7).
The Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) study initiated in 2013 revealed that the rate of incident carbapenem-resistant Enterobacteriaceae clinical cultures in government hospitals in Singapore increased during 2011–2013 and plateaued thereafter (8). The number of cases of hypervirulent K. pneumoniae has increased in the past 3 decades in parts of Asia, and likewise, the number of cases of monomicrobial Klebsiella-induced liver abscesses has also increased (9,10).
The prevalence of antimicrobial resistance among hypervirulent K. pneumoniae isolates is rare compared with that of standard isolates (11,12); hypervirulent K. pneumoniae and CRKP seem to have their own particular reservoirs and remain mostly segregated from each other. However, hypervirulent K. pneumoniae and CRKP isolates can converge in the same organism, leading to the emergence of superbugs resistant to antimicrobial drugs of even the last line of treatment that are capable of infecting healthy persons. This emergence has already been reported in China, Brazil, and the United Kingdom (13–15). The fatal outbreak that occurred in a hospital in China in 2016 was caused by a carbapenem-resistant hypervirulent K. pneumoniae strain that had acquired a virulence plasmid by a classic sequence type (ST) 11 strain (16). In a study of a collection of >2,200 K. pneumoniae genomes, distinct evolutionary patterns of horizontal gene transfer were observed in MDR isolates versus hypervirulent isolates (17). The authors of that study postulated that hypervirulent clones might be subject to some sort of constraint against horizontal gene transfer and show more conserved pangenomic diversity than MDR clones. If that hypothesis is correct, MDR clones acquiring virulence genes or K. pneumoniae virulence plasmids would be more likely than hypervirulent clones acquiring MDR genes. To investigate this hypothesis, we searched for hypervirulent isolates among 556 CRKP isolates collected at public hospitals of Singapore.
Bacterial Isolates and Microbiologic Methods
During 2010–2015, all microbiology laboratories in Singapore had been mandated to submit their carbapenem-resistant Enterobacteriaceae isolates to the National Public Health Laboratory of Singapore. Using this library, we collected isolates from the CaPES study. We performed species identification, assessed carbapenem resistance, and determined carbapenemase genes as previously described (8).
Whole-Genome Sequencing and Data Analysis
We performed whole-genome sequencing using the MiSeq platform (Illumina, ) as previously described (18). In addition, we sequenced the complete genomes of 5 isolates from 3 patients, obtaining long reads using the GridION X5 system (Oxford Nanopore Technologies, ) to close the gaps. We de novo assembled the Illumina sequence reads using SPAdes 3.11.1 (19) and completed genome assembly using a combination of Illumina and Oxford Nanopore Technologies data with the hybrid assembler Unicycler version 0.4.7 (Appendix Table) (20). We deposited whole-genome sequencing data in GenBank (BioProject numbers PRJNA342893, PRJNA557813, and PRJNA591409). We screened genome assemblies for virulence loci and K. pneumoniae virulence plasmid–associated loci using kleborate (21–23). We resolved missing loci and ambiguous alleles by mapping short reads to reference sequences using breseq (24) and screened assemblies for antimicrobial resistance genes using ResFinder 3.1 (25) and CARD (26). We resolved any discrepancies between these 2 gene identifiers by searching blastp () using translated gene sequences. We identified plasmid replicons in all completely sequenced genomes using PlasmidFinder with default settings (27). For all isolates, we performed core-genome single-nucleotide polymorphism (SNP) analysis against reference genome SGH10 chromosome (GenBank accession no. CP025080) using Parsnp 1.2 (28). In plasmid analyses, we generated alignments by mapping assemblies to reference plasmids using bowtie2 (29) on the REALPHY server (30). We inferred approximate maximum-likelihood phylogenetic trees using FastTree 2 (31) and screened completed assemblies for origin of transfer (oriT) sites and other transfer-related modules using oriTfinder (32).
Determining Hypermucoviscosity
Mouse Infection
We infected female 7–8-week-old C57BL/6J mice (InVivos, ) with 1 × 105 CFU of bacteria diluted in 100 μL phosphate-buffered saline through the intraperitoneal route and assessed for death every 8–16 h. Animal experiments were approved under protocol R18–0252 by the National University of Singapore Institutional Animal Care and Use Committee in accordance with the National Advisory Committee for Laboratory Animal Research guidelines.
Conjugation Experiments
We measured the transmissibility of the blaKPC-2–carrying plasmid using a previously described method (35). In this experiment, carbapenem-resistant hypervirulent K. pneumoniae isolates were the donors and a kanamycin-resistant Escherichia coli MG1655 mutant SLC568 strain (36) was the recipient. We carried out conjugation on 0.22-µm nitrocellulose filters with donors and recipients incubated at a 1:1 ratio on lysogeny broth (LB) agar plates for 4 h at 37°C. We enumerated transconjugants on LB agar plates containing carbenicillin (100 µg/mL) and kanamycin (50 µg/mL) and recipients on LB agar plates containing kanamycin only. We confirmed transfer of the blaKPC-2 gene by PCR.
Antimicrobial Susceptibility Testing
Statistical Methods
We performed statistical analyses using GraphPad Prism version 8 (). We compared samples using the unpaired t-test with Welch correction.
Discovery of Hypervirulent Features of CRKP
We retrieved 1,312 carbapenem-resistant Enterobacteriaceae collected from 6 public hospitals in Singapore during 2010–2015 through the CaPES program and National Public Health Laboratory of Singapore; 1,251 isolates were whole-genome sequenced with Illumina technology, and 556 isolates were K. pneumoniae. We searched K. pneumoniae isolate genomes for the presence of K. pneumoniae virulence plasmid–associated virulence determinants, rmpA, rmpA2, iro (the salmochelin locus), and iuc (the aerobactin locus) by using kleborate. We identified 18 isolates (originating from just 7 patients) harboring all of these loci, and 14 of these isolates came from the same 3 patients. We screened the genome assemblies of these 18 isolates for virulence features and compared the characteristics of these isolates with those of 2 known hypervirulent strains, SGH10 (serotype K1 liver abscess–associated isolate from Singapore) (6,39) and CG43 (serotype K2 clinical isolate from Taiwan) (40). We then performed a phylogenetic analysis of the core genomes of all these isolates.
The differences found among isolates from the same patient were small (0–15 SNPs) (Figure 1, panel A), suggesting that patients with multiple isolates were infected with a single strain. All isolates from patients A2, A4, and A6 were ST23 and serotype K1 (same as SGH10) and, except for ENT1256, carried the same virulence loci as SGH10 (Table 1); ENT1256 had a different allele for rmpA2. The core genomes of these isolates were also similar to SGH10, as shown by the phylogenetic analysis (Figure 1, panel A). However, the differences between isolates from patients A2, A4, and A6 were much greater (200–300 SNPs) than the differences between isolates from patient A2 (15 SNPs), indicating that the bacteria from these patients were unlikely to have originated from the same strain. The isolates from patients A14 and A15 were the same serotype (K2) but different sequence types (Table 1), and their core genomes contained many differences (>20,000 SNPs). The isolates from patient A14 were phylogenetically close to the other hypervirulent strain, CG43 (Figure 1, panel A); however, unlike CG43, which carried no yersiniabactin (ybt) and colibactin (clb) loci, 2 A14 isolates (ENT1192 and ENT1988) had a ybt 9 locus on integrative conjugative element K. pneumoniae 3 (Table 1). The isolates from the remaining 2 patients (A8 and A12) were the same serotype (K20) but had different sequence types and many core genome differences (>20,000 SNPs). ENT1332 had an unknown ybt locus, and ENT1381 had a ybt 9 locus on integrative conjugative element K. pneumoniae 3. Both of these isolates did not have the clb locus. All isolates had hypervirulence backgrounds.
We also performed a whole-genome phylogenetic analysis on the K. pneumoniae virulence plasmid carried in the first isolates obtained from all patients, using the K. pneumoniae virulence plasmid sequence from SGH10 (pSGH10) as reference. The K. pneumoniae virulence plasmids appeared to form 2 separate clades, 1 for all K1 isolates and the other for K2 and K20 isolates (Figure 1, panel B).
The virulence potentials of all isolates were high (Table 1); the virulence scores predicted by kleborate for all isolates (>4) were higher than the score (3) predicted for the hypervirulent K2 reference strain CG43. Hypervirulent K. pneumoniae isolates are generally defined as carrying K. pneumoniae virulence plasmid–associated loci and having a hypermucoviscous phenotype (including a positive string test result), which is dependent on regulator RmpA (40,41). We measured the hypermucoviscosity of the first isolates from all patients using both the string test and a centrifugation assay; 6 of 7 isolates formed strings, and 5 of 7 isolates were hypermucoviscous according to the centrifugation assay (Figure 2, panel A). Only ENT596 was negative by both tests.
The virulence potential of the carbapenem-resistant hypervirulent K. pneumoniae isolates was further determined in an intraperitoneal mouse infection model. The first isolates from 6 of 7 patients killed >50% of the infected mice within 96 hours; only ENT596 (like control isolate ENT495, a CRKP strain carrying pKPC2 but not hypervirulent) did not kill any mice (Figure 2, panel B). Serotype K1 isolates were the most virulent, and serotype K20 isolates took longer to kill. The virulence of ENT596 did not correlate with its predicted score; this isolate demonstrated concurrent loss of hypermucoviscosity and virulence in mice (Table 1; Figure 2), which might have been caused by the loss of expression of the virulence genes. The average length of hospitalization of the 7 patients harboring these isolates was 97 days, which was much longer than the average length of stay for 249 patients with carbapenem-resistant Enterobacteriaceae infection (38 days) (8). Taking into account all our evidence, we conclude that the isolates from 6 of 7 patients (A2, A4, A6, A8, A12, and A14) are phenotypically hypervirulent, and the isolates from patient A15 are phenotypically nonhypervirulent (although these isolates have a hypervirulent genetic background).
Highly Conserved Plasmid Harboring blaKPC-2 on All Carbapenem-Resistant Hypervirulent K. pneumoniae Isolates
We screened the assemblies of all 18 carbapenem-resistant hypervirulent K. pneumoniae isolates for acquired antimicrobial resistance genes using ResFinder (25) and CARD (26). Except for endogenous penicillin resistance, hypervirulent K. pneumoniae isolates are generally considered to be susceptible to antimicrobial drugs. Our search results showed that SGH10 harbored the β-lactam resistance gene blaSHV-11 and resistance genes against fluoroquinolones (oqxA and oqxB) and fosfomycins (fosA6) (Table 2). In total, 17 of 18 isolates carried these 3 resistance genes; 1 isolate from patient A15 (ENT686) did not have the oqxA and oqxB genes, and these genes were also not detectable by PCR. All isolates carried an identical set of the following 4 genes: blaKPC-2, blaTEM-1A, blaTEM-1B, and mph(A). In the 5 completely sequenced genomes (ENT494, ENT646, and ENT1734 [patient A2]; ENT1192 [patient A14]; and ENT607 [patient A15]; Appendix Table), we located these 4 genes on a 71,861-bp plasmid, which we named pKPC2. The sequences of the pKPC2s in the 4 isolates from patients A2 and A14 were identical. The pKPC2 in ENT607 from patient A15 had only 1-bp difference.
A blastn search for pKPC2 revealed that the highest hit was a plasmid from Salmonella enterica (pSA20021456.2, GenBank accession no. CP030221), which covered 74% of pKPC2 with >99% identity (Figure 3). No attribute information was available for the Salmonella strain, and pSA20021456.2 carries no antimicrobial drug resistance genes. No incompatibility groups were detected on either plasmid by PlasmidFinder.
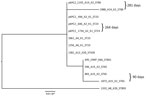
We performed a phylogenetic analysis of the pKPC2 sequences carried in the first isolates and other select isolates from all 7 patients, including the pKPC2 from ENT494 (i.e., pKPC2_494) as reference (Figure 4). This analysis revealed that all isolates carried sequences almost identical to pKPC2_494 (coverage and identity >99%). Because all isolates carried the 4-gene set, they probably had a plasmid closely related to pKPC2_494. Three patients had multiple isolates with pKPC2. The clinical records show long time-interval gaps of antimicrobial drug nonexposure between some isolates. The comparison also shows that the pKPC2-related plasmids were remarkably stable; they were maintained in bacteria with few changes for 90–281 days in the patients not undergoing antimicrobial drug treatment (Figure 4).
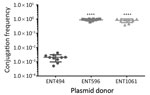
Besides resistance genes, pKPC2 had a complete set of conjugative machinery with all 4 essential modules (oriT, relaxase, type IV coupling protein, and a type IV secretion system cluster), suggesting the plasmid is self-transmissible. The Salmonella plasmid pSA20021456.2 has a relaxase, type IV coupling protein, and type IV secretion system cluster similar to pKPC2 but no oriT site. We selected 3 isolates that had only 1 antimicrobial drug resistance plasmid to assess the transmissibility of pKPC2. We performed filter mating on LB agar using the kanamycin-resistant E. coli strain SLC568 as the recipient. After 4 hours of incubation, ≈80% of recipients (≈0.8 × 100 transconjugate/recipient) had received pKPC2 from ENT596 and ENT1061 (Figure 5). The conjugation frequency was ≈0.2% (≈2.0 × 10–4 transconjugate/recipient) when ENT494 was the donor. We confirmed the transconjugants acquired the blaKPC-2 gene by PCR (using 10 colonies for each donor). K serotype, sequence type, and hypermucoviscosity of donors could not explain the observed differences in conjugation efficiency.
To assess the role of pKPC2 in conferring antimicrobial drug resistance to host strains, we measured the MICs of antimicrobial drugs against the 3 isolates from patient A2 and compared with SGH10 (Table 3). SGH10 was resistant to penicillins and fosfomycin but susceptible to the 2 fluroquinolones tested, even though the strain carried the fluroquinolone efflux pump genes oqxA and oqxB (Table 2). This finding is consistent with the low oqxB expression seen for most K. pneumoniae strains (42). SGH10 was susceptible to cephems and carbapenems. In contrast, all 3 isolates from patient A2 were resistant to ceftriaxone, imipenem, and meropenem (Table 3), most likely because of the presence of pKPC2. These data show that pKPC2 is a highly transmissible plasmid that confers resistance to all 3 types of β-lactams.
Within-Patient Microevolution of Carbapenem-Resistant Hypervirulent K. pneumoniae Isolates
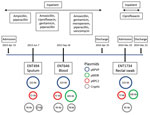
Using multiple isolates from the same patient, we set out to determine the changes that occurred in the genome of 1 carbapenem-resistant hypervirulent K. pneumoniae population over the course of an infection. This analysis enabled us to track the carriage of genes conferring carbapenem resistance and MDR in the bacteria versus antimicrobial drug exposure over time. With the available clinical data from patient A2, we reconstructed a timeline of the evolution of the 3 isolates from this patient, showing their plasmid content and antimicrobial drug exposure (Figure 6).
In addition to the chromosome, K. pneumoniae virulence plasmid, and pKPC2, all 3 isolates from patient A2 carried 2 or 3 additional plasmids (Appendix Table). Isolates were of the same infecting strain but lost or gained MDR plasmids over time. The K. pneumoniae virulence plasmid and pKPC2 were stable, showing few changes over a year. Treatment with gentamicin and ciprofloxacin correlated with the appearance of an MDR plasmid in ENT646 that encoded resistance genes to both classes of antimicrobial drugs. This 165-kb MDR plasmid (named pMDR646) carried aac(6')-lb-cr, blaOXA-1, qnrB1, catB3, and dfrA14. In the third isolate from patient A2 (ENT1734), the MDR plasmid downsized to 100 kb (pMDR1734) and carried only aac(6')-lb-cr, blaOXA-1, and catB3. The downsizing appeared to be the result of extensive deletion and reorganization (Appendix Figure). Antimicrobial susceptibility testing data also showed that this bacterium was resistant to fewer antimicrobial drugs; isolate ENT646 was resistant to 7 classes of antimicrobial drugs, and ENT1734 was resistant to 6 classes of drugs (Table 3). Both ENT646 and ENT1734 carried the broad-range resistance gene aac(6')-lb-cr, which confers resistance against aminoglycoside and fluoroquinolone antimicrobial drugs. This gene was likely responsible for the observed resistance to kanamycin, and qnrB1 (in ENT646) was likely responsible for resistance to ciprofloxacin (Table 3). Both isolates were also resistant to sulfamethoxazole, which we could not attribute to any resistance gene or mutation. All 3 isolates carried a 95-kb plasmid with unknown virulence or resistance attributes (Figure 6). The sizes of the chromosome and K. pneumoniae virulence plasmid gradually increased over time, mainly through the acquisition of mobile genetic elements.
Nine isolates were collected from patient A15 at various time points and from different anatomic sites (Table 2); all were identical in their core genomes (0 SNP differences). Two A15 isolates acquired antimicrobial drug resistance genes in addition to those on pKPC2; 1 of the first isolates, ENT607, had aadA1, mph(A), cmlA1, arr-2, and sul1, which were all on a 105-kb plasmid (named pMDR607). This plasmid appeared unstable and was absent in later isolates. The last isolate, ENT1077, had acquired tetracycline and trimethoprim resistance genes not seen in the other isolates.
We report the coexistence of hypervirulence and carbapenem resistance within the same K. pneumoniae isolates in Singapore. These isolates dated back to 2013; however, their existence could have occurred even earlier because collection started in 2010. All carbapenem-resistant hypervirulent K. pneumoniae isolates in this study harbored a blaKPC-2 gene. In studies conducted in China, most patients infected with carbapenem-resistant hypervirulent K. pneumoniae also carried the blaKPC-2 gene (16,43–45). We and others have described hypervirulent K. pneumoniae in community-acquired liver abscesses in Singapore, where most isolates are capsular serotypes K1 and K2 (6,46). Most (>80%) liver abscess isolates are estimated to belong to sublineage CG23-I (includes reference strain SGH10) (39). The carbapenem-resistant hypervirulent K. pneumoniae isolates from 5 of 7 patients in this study were K1 or K2 serotype and 6 of 7 isolates were highly virulent, as predicted. Our results suggest that pKPC2 can be stably maintained in a hypervirulent K. pneumoniae bacterial host. Thus, the possible dissemination of the blaKPC-2 gene to hypervirulent strains present in a carriage state within communities is a concern.
In a study in China, 1,838 isolates were analyzed, and 21 carbapenem-resistant hypervirulent K. pneumoniae isolates were found and classified as ST11, ST65, ST268, ST595, and ST692 (44). In another report, 5 ST11 CRKP isolates were documented as having acquired K. pneumoniae virulence plasmids (16). In these reports, a K. pneumoniae virulence plasmid or parts of one was probably co-opted into the prevalent CRKP strain in that region, which was an ST11 strain carrying the blaKPC-2 gene. According to our previous study, only 5% of CRKP isolates in Singapore were ST11 (8). However, in another study, 7 carbapenem-resistant hypervirulent K. pneumoniae isolates were identified, 4 of which were ST23 and serotype K1 (47). In 2 case studies, ST23 (hypervirulent) isolates were reported to have acquired carbapenem-resistance genes (14,48). All our isolates in this investigation had the hypervirulence background and acquired carbapenem-resistance and MDR genes; the MDR genes appeared to move in and out of the parental strain over time within the patient. Our observations are consistent with earlier reports in China involving ST65 isolates, which were likely hypervirulent, indicating that hypervirulent isolates can acquire antimicrobial drug resistance. Therefore, whether a superbug is more likely to arise from a carbapenem-resistant isolate acquiring a K. pneumoniae virulence plasmid or some of its genes versus a hypervirulent isolate acquiring MDR genes is unclear. Both ways of exchange appear possible, perhaps depending on the prevalence of the circulating sequence type in a particular setting and in vivo selection pressures.
We also show that, although isolates from different patients were similar in terms of the virulence and carbapenem-resistance plasmids, these isolates do not arise from transmission events. For patients with multiple isolates, the core genomes were highly conserved, suggesting a single infecting isolate gained or lost various antimicrobial drug resistance genes over time. For patient A2, our longitudinal data suggest that the use of distinct antimicrobial drugs drove the acquisition of resistance, though we have no direct proof of cause and effect. Of note, in patient A15, the rectal isolate had additional antimicrobial drug resistance, perhaps reflecting that the colon might serve as a reservoir where genetic exchange can take place. Six of the 18 isolates, including the last acquired isolates from the 3 patients with multiple isolates, were collected from feces or rectal swab specimens, suggesting that the intestines are a likely reservoir for persistent carriage. Last, we demonstrated as a proof-of-principle that pKPC2 can be transferred from the K. pneumoniae isolates to E. coli in vitro with high efficiency. Determining the particular characteristics of pKPC2 that make it so transmissible and stable, particularly during interactions with hypervirulent isolates, and how pKPC2 is acquired by the recipient is essential.
Therefore, this report serves to alert infectious disease clinicians to the possible presence of hypervirulence in MDR or carbapenem-resistant colonizing isolates; patients harboring these isolates are at risk of developing invasive infections. With timely (rather than retrospective) whole-genome sequencing of bacteria, identifying patients harboring hypervirulent and multidrug or carbapenem-resistant isolates at high risk for death should be possible. These patients can be selected for appropriate infection control measures, treatment, and close monitoring. Developing strategies to decolonize the gastrointestinal tracts of patients with such isolates would help minimize the release of these potential superbugs into the community.
Mr. Chen is a research associate at the Yong Loo Lin School of Medicine at National University of Singapore, Singapore. His main research interests are regulation of virulence and antimicrobial drug resistance in bacterial pathogens, including K. pneumoniae.
Acknowledgments
We thank the CaPES study group for their support. This group included Benjamin Cherng, Deepak Rama Narayana, Douglas Chan Su Gin, De Partha Pratim, Hsu Li Yang, Indumathi Venkatachalam, Jeanette Teo, Kalisvar Marimuthu, Koh Tse Hsien, Nancy Tee, Nares Smitasin, Ng Oon Tek, Ooi Say Tat, Raymond Fong, Raymond Lin Tzer Pin, Surinder Kaur Pada, Tan Thean Yen, and Thoon Koh Cheng. We thank Ewan M. Harrison for his critical reading of the manuscript.
This work was supported by the National University of Singapore, Yong Loo Lin School of Medicine Aspiration Fund (NUHSRO/2014/068/AF New Idea/03), and National Medical Research Council (NMRC) collaborative grant Collaborative Solutions Targeting Antimicrobial Resistance Threats in Health Systems (CoSTAR-HS, HS/ARGSeedGrant/2017/03) to Y.-H.G. Additional grant support was provided by CoSTAR-HS (NMRC CGAug16C005), NMRC Clinician Scientist Award (NMRC/CSA-INV/0007/2016), and NMRC Clinician Scientist Award (MOH-000276).
References
- Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15.
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al.; EuSCAPE Working Group; ESGEM Study Group. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29.
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–96.
- Government of the United Kingdom. Review on antimicrobial resistance. tackling drug-resistant infections globally: an overview of our work. 2016 Mar [cited 2019 Aug 22].
- World Health Organization. Worldwide country situation analysis: response to antimicrobial resistance. 2015 Apr 29 [cited 2019 Aug 22].
- Lee IR, Molton JS, Wyres KL, Gorrie C, Wong J, Hoh CH, et al. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep. 2016;6:29316.
- Chew KL, Lin RTP, Teo JWP. Klebsiella pneumoniae in Singapore: hypervirulent infections and the carbapenemase threat. Front Cell Infect Microbiol. 2017;7:515.
- Marimuthu K, Venkatachalam I, Khong WX, Koh TH, Cherng BPZ, Van La M, et al.; Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group. Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae among adult inpatients in Singapore. Clin Infect Dis. 2017;64(suppl_2):S68–75.
- Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12:881–7.
- Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32:
e00001-19 . - Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895.
- Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–61.
- Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483.
- Roulston KJ, Bharucha T, Turton JF, Hopkins KL, Mack DJF. A case of NDM-carbapenemase-producing hypervirulent Klebsiella pneumoniae sequence type 23 from the UK. JMM Case Rep. 2018;5:
e005130 . - Andrade LN, Vitali L, Gaspar GG, Bellissimo-Rodrigues F, Martinez R, Darini AL. Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol. 2014;52:2530–5.
- Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46.
- Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:
e1008114 . - Khong WX, Marimuthu K, Teo J, Ding Y, Xia E, Lee JJ, et al.; Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group. Tracking inter-institutional spread of NDM and identification of a novel NDM-positive plasmid, pSg1-NDM, using next-generation sequencing approaches. J Antimicrob Chemother. 2016;71:3081–9.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . - Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4.
- Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10:77.
- Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:
e000102 . - Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–88.
- Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4.
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–73.
- Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903.
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
- Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 2014;31:1077–88.
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:
e9490 . - Li X, Xie Y, Liu M, Tai C, Sun J, Deng Z, et al. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018;46(W1):W229–34.
- Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705.
- Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol. 2003;185:788–800.
- Hardiman CA, Weingarten RA, Conlan S, Khil P, Dekker JP, Mathers AJ, et al. Horizontal transfer of carbapenemase-encoding plasmids and comparison with hospital epidemiology data. Antimicrob Agents Chemother. 2016;60:4910–9.
- Khetrapal V, Mehershahi K, Rafee S, Chen S, Lim CL, Chen SL. A set of powerful negative selection systems for unmodified Enterobacteriaceae. Nucleic Acids Res. 2015;43:
e83 . - Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically (M07-A10). Wayne (PA): The Institute; 2015.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing (M100-S28). Wayne (PA): The Institute; 2018.
- Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan YH, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9:2703.
- Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–98.
- Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62:1–6.
- Zheng JX, Lin ZW, Sun X, Lin WH, Chen Z, Wu Y, et al. Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7:139.
- Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–12.
- Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 Isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7:182.
- Lin YT, Siu LK, Lin JC, Chen TL, Tseng CP, Yeh KM, et al. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 2012;12:13.
- Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2015;60:709–11.
- Cejas D, Fernández Canigia L, Rincón Cruz G, Elena AX, Maldonado I, Gutkind GO, et al. First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J Clin Microbiol. 2014;52:3483–5.
- Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71:553–60.
Figures
Tables
Cite This ArticleOriginal Publication Date: 2/6/2020





































No hay comentarios:
Publicar un comentario