
Epigenetics: A New Tool for Forensic Detectives
 Thought LeadersDr Bruce R. McCordProfessor of ChemistryFlorida International University
Thought LeadersDr Bruce R. McCordProfessor of ChemistryFlorida International UniversityAn interview with Dr. Bruce R. McCord, conducted by Alina Shrourou, BSc
It has been announced that you will be presenting in the “Analytical Methods in Forensic Biology and DNA Analysis” symposium at Pittcon 2018. Why are bioanalytical methods important for forensics?
Developing analytical methods can help the triers of fact, judges and juries, better understand the events surrounding the circumstances of a crime. In forensic analysis, we are talking about determining the identity of unknown individuals through DNA and using DNA and other chemical signatures to determine and clarify the circumstances of the crime.
The advantage of modern analytical methods is their specificity and the ability to perform statistical analysis on resultant data. Examples include using laser induced fluorescence to detect trace levels of amplified DNA at a crime scene or using spectroscopic analysis to detect biological stains.
This improved precision and increased sensitivity that bioanalytical methods allow, mean that forensic scientists can help the jury reach a valid conclusion on what has happened.
What will be the main focus of your talk titled “Forensic Epigenetics, A Novel Method for Body Fluid Identification and Phenotyping”?
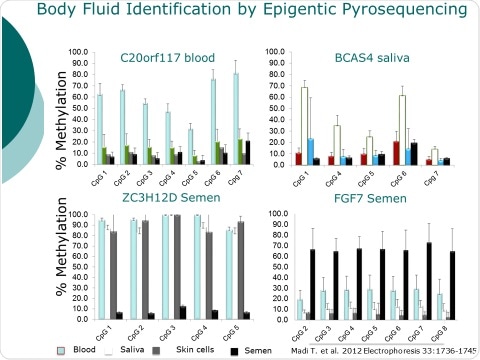
I will be presenting our work on age and tissue type. We will demonstrate how epigenetic DNA methylation permits the determination of body fluid type, suspect age, and other information from trace levels of samples left behind at crime scenes.
 © petarg/Shutterstock.com
© petarg/Shutterstock.comWhat is epigenetic methylation and how can it be detected?
Epigenetics involves changes in gene function that are not related to changes in the DNA sequence. It is well known that all human DNA is the same, and is comprised of the same nucleiotide bases. However, it is easy to see that cells are differentiated based on their application - skin, muscle, bone, etc. These differences are the result of molecular switches which determine gene expression.
One such switch involves differential methylation of cytosine – one of the four main bases found in DNA. It is common to find locations upstream of various genes known as CpG islands (a cytosine base immediately followed by a guanine) in which clusters of methylated cytosines occur. When the methylation level of a specific tissue is diagnostic, the procedure can be used to determine the cell type. Similarly, other locations in the genome can indicate a suspects age.
The presence of methylation at these locations can be detected in various ways. A common method is to use bisulfite modified PCR. This process chemically converts unmethylated GC base pairs to AT base pairs through chemical modification and PCR amplification.
Methylated cytosines are not converted. The result of this process is then easily read using DNA sequencing methods.
Please outline the sequencing methods involved with locating epigenetic loci.
We mainly use 3 methods to read the DNA and determine whether methylation is present at specific loci. Firstly, methylation array data helps us determine potential sites of interest in the entire human genome.
We then examine and score this data and look for specific CpGs in the array data which may indicate the presence of sequences of interest. We design specific primers to target regions of interest in the genome and use pyrosequencing to determine if these locations are probative. These locations are then probed using bisulfite modified PCR and pyrosequencing.
Lastly, because of the difference in melting temperature between GC and AT basepairs, we can also target locations using real time PCR with high resolution melt capability as long as large differences in methylation are present.
How can epigenetic methylation be used in the crime scene?
We use this procedure to determine trace levels of body fluids. This can be particularly important in situations involving crimes such as sexual assault or child abuse, where the suspect is present and finding their DNA would not be unexpected.
However, the type of DNA recovered - whether it is from a body fluid, compared to from skin cells for example, might indicate if a crime had in fact occurred. Similarly, if a number of suspects are identified, the determination of age of an unknown sample from a crime scene would be very important.
How can CpG loci identify the lifestyle choices of an individual and how valid are these assumptions?
Lifestyle choices such as smoking may alter the methylation level at certain CpGs. In many cases, just like tissue specific epigenetics, these loci are linked to specific genes which are activated by the need of the body to mediate the lifestyle effect. Validity can only be determined by determination of gene function and experiments with population samples, but this is an important area of research in many biomedical fields.
We are just beginning to examine this area, but we note previous research exists determining links between epigenetic markers and factors such as smoking.
What data will you be presenting during your talk and how does this compare to other research in the field?
While all forensic research deals with human identity, phenotyping and use in assessing the biological information left by suspects at a crime scene, our focus is different from most other research in the field as we are targeting markers of interest in forensic science. Others in this field are more concerned with biomedical applications.
As epigenetic loci originate with DNA, we can use the power of being at the root of the gene expression tree. By performing bisulfite modification prior to amplification, we have an advantage when it comes to DNA phenotyping as this means our process is resistant to inhibition and factors which may intervene with gene expression. Other methods using RNA or proteomics as used in traditional forensics research, may be subject to variations in gene expression which require correction.
Thus, we envision that a portion of the forensic DNA sample can be diverted for age and tissue determination.
Are there any other advantages of using epigenetic loci in body fluid identification over other methods?
It is important to us to ensure that our interfaces work well with traditional forensic techniques, so that it is more easily implemented in a laboratory. Therefore, perhaps the biggest advantage to epigenetic analysis is that the extraction of the sample and recovery of the DNA is the same with standard DNA typing and forensic epigenetics.
Additionally, as methylation involves covalent bonding, this method is extremely stable – samples over 20 years old can be analysed this way!
Where do you see epigenetics developing in forensic biology?
I see epigenetics developing as a tool to compliment standard forensic DNA typing, and our work is likely to contribute to this development. I think that future methods involving massively parallel sequencing (MPS), also known as next generation sequencing (NGS), will include procedures for simultaneous genotyping and epigenetic analysis.
As far as advancing the field it is too soon to know. However, we believe that epigenetics can be an important tool in trace serology and in forensic investigations of individuals not yet in a database due to its capability to detect age and certain phenotypic information.
Where can readers find more information?
The paper relating to our research can be found here:
Madi, T., Balamurugan, K., Bombardi, R., Duncan, G., & McCord, B. (2012). The determination of tissue‐specific DNA methylation patterns in forensic biofluids using bisulfite modification and pyrosequencing. Electrophoresis, 33: 1736-1745.
I would like to acknowledge the students and collaborators who worked on the project cited, including Tania Madi, Joana Antunes, Debora Silva, Hussain Alghanim (FIU) Kuppareddi Balamurugan (U. Southern Mississippi) George Duncan (Broward Sheriff’s Office) and Clarice Alho (Pontifico Catholic University of Rio Grande Do Sul, Brazil). I would also like to thank the National Institute of Justice for providing the funding for the work, as well as the technical assistance from Qaigen.
There are also a number of other papers you can refer to for more information:
- Alghanim, H., Antunes, J., Silva, D. S. B. S., Alho, C. S., Balamurugan, K., & McCord, B. (2017). Detection and evaluation of DNA methylation markers found at SCGN and KLF14 loci to estimate human age. Forensic Science International: Genetics, 31: 81-88.
- Antunes, J., Silva, D. S., Balamurugan, K., Duncan, G., Alho, C. S., & McCord, B. (2016). Forensic discrimination of vaginal epithelia by DNA methylation analysis through pyrosequencing. Electrophoresis, 37: 2751-2758.
- Silva, D. S., Antunes, J., Balamurugan, K., Duncan, G., Alho, C. S., & McCord, B. (2016). Developmental validation studies of epigenetic DNA methylation markers for the detection of blood, semen and saliva samples. Forensic Science International: Genetics, 23: 55-63.
- Antunes, J., Silva, D. S., Balamurugan, K., Duncan, G., Alho, C. S., & McCord, B. (2016). High-resolution melt analysis of DNA methylation to discriminate semen in biological stains. Analytical biochemistry, 494: 40-45.
- Soares Bispo Santos Silva, D., Antunes, J., Balamurugan, K., Duncan, G., Sampaio Alho, C., & McCord, B. (2015). Evaluation of DNA methylation markers and their potential to predict human aging. Electrophoresis, 36: 1775-1780.
About Dr Bruce McCord
 Bruce R. McCord is currently Professor of Analytical and Forensic Chemistry at Florida International University. He received a BSc in Chemistry with honors from the College of William and Mary and his PhD in Analytical Chemistry from the University of Wisconsin- Madison.
Bruce R. McCord is currently Professor of Analytical and Forensic Chemistry at Florida International University. He received a BSc in Chemistry with honors from the College of William and Mary and his PhD in Analytical Chemistry from the University of Wisconsin- Madison.His present research interests involve the development of applications in forensic genomics, epigenetics, microfluidics and nanoscale sensing. Major research areas include development of improved techniques for genetic analysis using capillary and microfluidic electrophoresis, DNA sequencing and QPCR.
He also has research efforts in toxicological applications of mass spectrometry, surface enhanced Raman spectroscopy and microfluidics.
He currently serves as Deputy Editor for the journal Electrophoresis and is a member of the editorial boards for the Journal of Forensic Science and the Journal of Forensic Chemistry. He has published over 100 peer reviewed articles and 12 book chapters and has graduated 20 PhD students, 15 MS students and supervised 12 post-doctoral researchers. In 2008 he received the Paul Kirk Award from the AAFS based on his contributions to the field of criminalistics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.





















.png)












No hay comentarios:
Publicar un comentario