
Investigating new cancer therapy candidates with live-cell imaging
 insights from industryProfessor Lawrence KwongAssistant Professor, Department of
insights from industryProfessor Lawrence KwongAssistant Professor, Department ofTranslational Molecular Pathology, Universityof Texas MD Anderson Cancer Center
An interview with Prof Lawrence Kwong, conducted by James Ives, MPsych
Please give an overview of your research into cancer and the mechanisms that you and your research team are looking to target with novel therapy candidates.
There are two main things that we've worked on in the lab as regards to drug studies. One is identifying new combinations of drugs, and working on why a combination of drugs should work better than each alone.
In the past, we've done this to identify the MEK + CDK4/6 combination for melanoma, we're hoping to use the same type of principle to move into the cholangiocarcinoma stage, cholangiocarcinoma is a bile duct cancer that's fairly rare in the west but it's extremely deadly.
In terms of survivability, it's almost right up there with pancreatic cancer, so we have a dire need of new therapies. We're testing now, for example, several different combinations, which include proprietary drugs evolved here MD Anderson, so we're really interested in attacking from multiple different angles and in ways that are novel.
How do your drug resistance studies in human samples help inform mechanistic insights of how drug resistance occurs?
We looked at human melanoma samples that had drug resistance to the combination that I mentioned previously, MEK + CDK4/6 inhibitors, and we identified a mutation in one patient. It was a fairly common mutation in cancer in general but, in the context of melanoma, hasn't really been studied well and good downstream targets hadn’t been identified.
The gene itself, the protein itself, is targetable, but there are some issues with moving that drug class into the clinic. Our idea was to move farther downstream, we conducted some unbiased protein array assays and identified a drug target that was uncommon for the mutation driving the resistance, and we modeled this in cell lines.
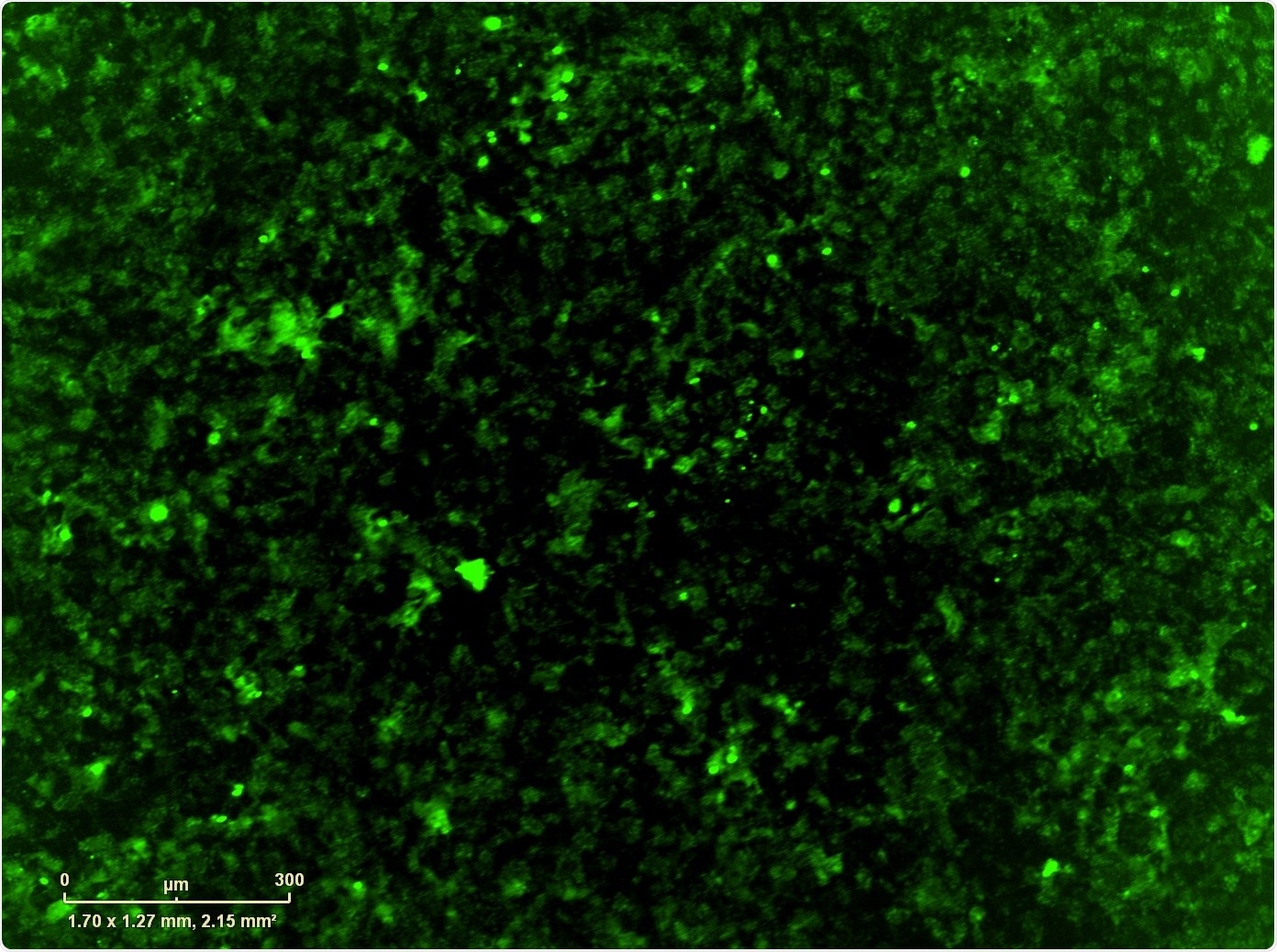
For this, we used Essen BioScience’s IncuCyte® Live-Cell Analysis System. Once we identified the counter-resistance drug, we combined it with the MEK+CDK combination which, if we didn't have the IncuCyte, it would have taken us a very long time to do every single combination that we wanted to do: we had over-expressed the resistance mutation, but also wild type version of the gene and the control GFP, so we had a number of different combinations in multiple cell lines that we wanted to test.
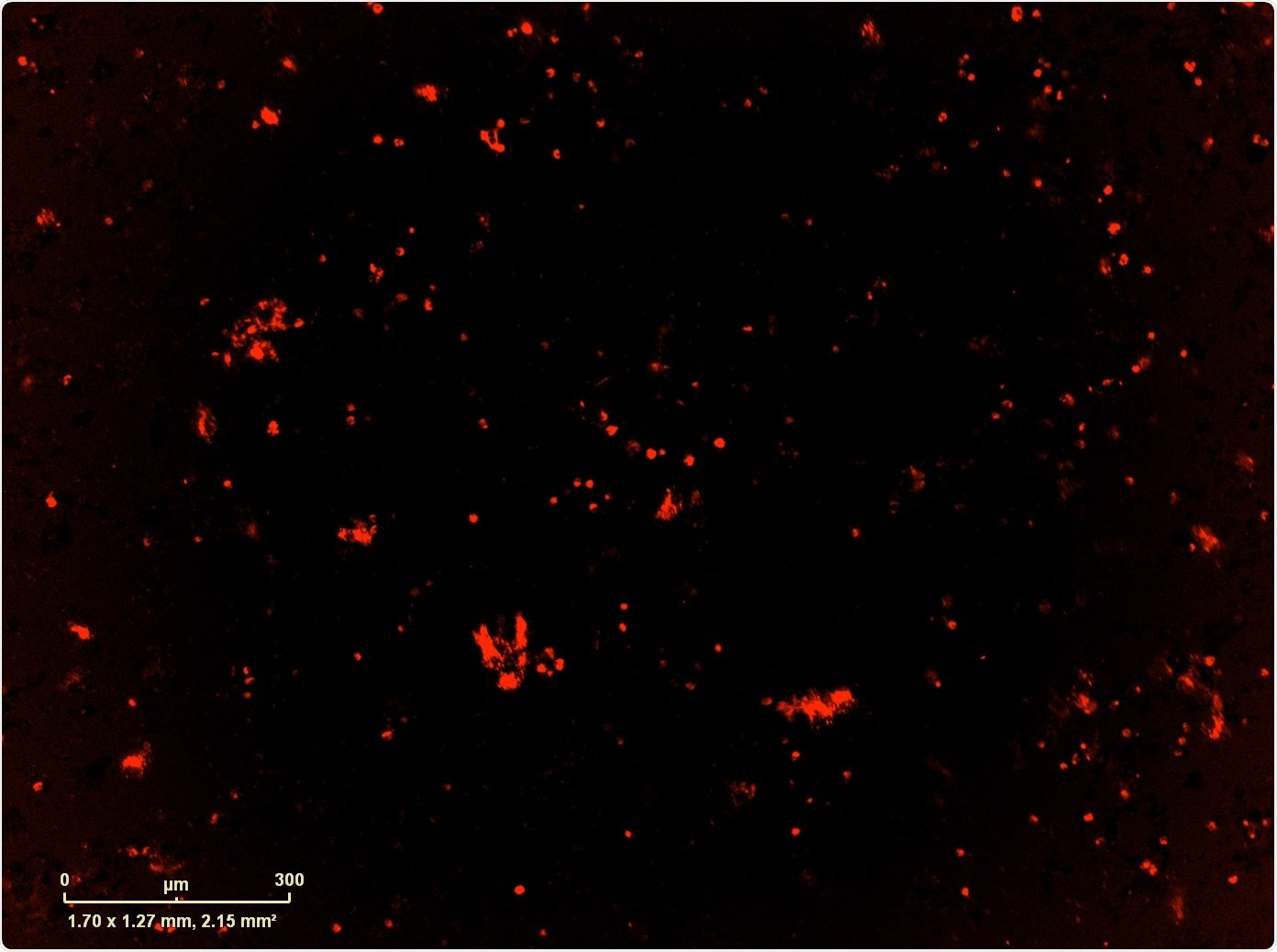
The IncuCyte not only allowed us to do that, but we also used it to image apoptosis and proliferation, which gave us some mechanistic components as well.
How does the dose of a treatment affect the resistance of cancer cells to that treatment? What research have you conducted into dose resistance of drug candidates in human samples?
That's a great question, in this study we conducted an 8-point dose curve for everything, every combination and the single drugs. The throughput was really a huge benefit. With the dosing that we used, essentially, we saw resistance across the board. No matter how high we went with the drug dose, we still saw resistance.
So at least in vitro we felt that we needed something really strong as the third drug to come in, and we feel like the drug and the drug target that we chose is very good.
The particular drug that we chose is one of the first and only drugs that came out for this target, we're waiting for second generation drugs to come out, but we're pretty happy with how it works. So, we didn't have to go very high with the drug dose to overcome the resistance to some degree.
What research are you and your group conducting to investigate cancer biomarkers and novel treatments that target these genes?
We are going to do a couple different things in the melanoma space as a follow up to this paper. What we're planning to do is to go through the archives of samples, knowing the samples that we have at MD Anderson which are many hundreds, and do DNA analysis to see how often we see the mutation we found and once we see them, we can look for this downstream marker which also serves as the drug resistance target.
For the cholangiocarcinoma research we're doing, as I mentioned it's a fairly rare cancer, but we found that we had 90 surgical samples at MD Anderson to work with and generate a tissue microarray and, hopefully, do some biomarker analysis there. We are also interested in developing novel drug combinations including some of the proprietary drugs that we have here.
How does live-cell imaging inform your research? How do you use the IncuCyte system?
I think the thing that we most like about the system has to do with the way that I approach science in the lab, which is that I'm kind of a perfectionist and I also second guess my own results all the time.
I always think about whether this or that could have gone wrong, whether it’s possible that we had uneven plating of the cells, whether it’s possible we had contamination of the well and other potential confounding variables.
These kind of things are very difficult to control for with, for example, a crystal violet assay or a cell titer glo assay, but the IncuCyte removed those fears we have about these problems for the most part.
We can go back into the data and see whether there was uneven plating, whether there was some weird clumping of cells in one well, and if so, we can look at what happens if we take that well out.
Then we can decide whether we need to repeat a well, or a cell line. We can see if there was contamination, which may not be so easily picked up if you had to go through every single well under a microscope yourself. Having the confidence in the results is huge aspect of it.
What methods were you previously using to complete your research? What advantages are there to using live-cell imaging with the IncuCyte system?
Primarily we were working with end-point analysis. We would take the cells after four days and we would stain them with crystal violet or cell titer glo, using a plate reader, which is very standard.
Those all have their own unique problems, especially that you can't do multiple analyses like the apoptosis analysis at the same time, which you can with IncuCyte.
There is also then the need to run a control. The way that we used to control for loading is to plate a replicate plate and add crystal violet to that after 24 hours with the hope that it's a rough estimate of how well you plated your actual wells.
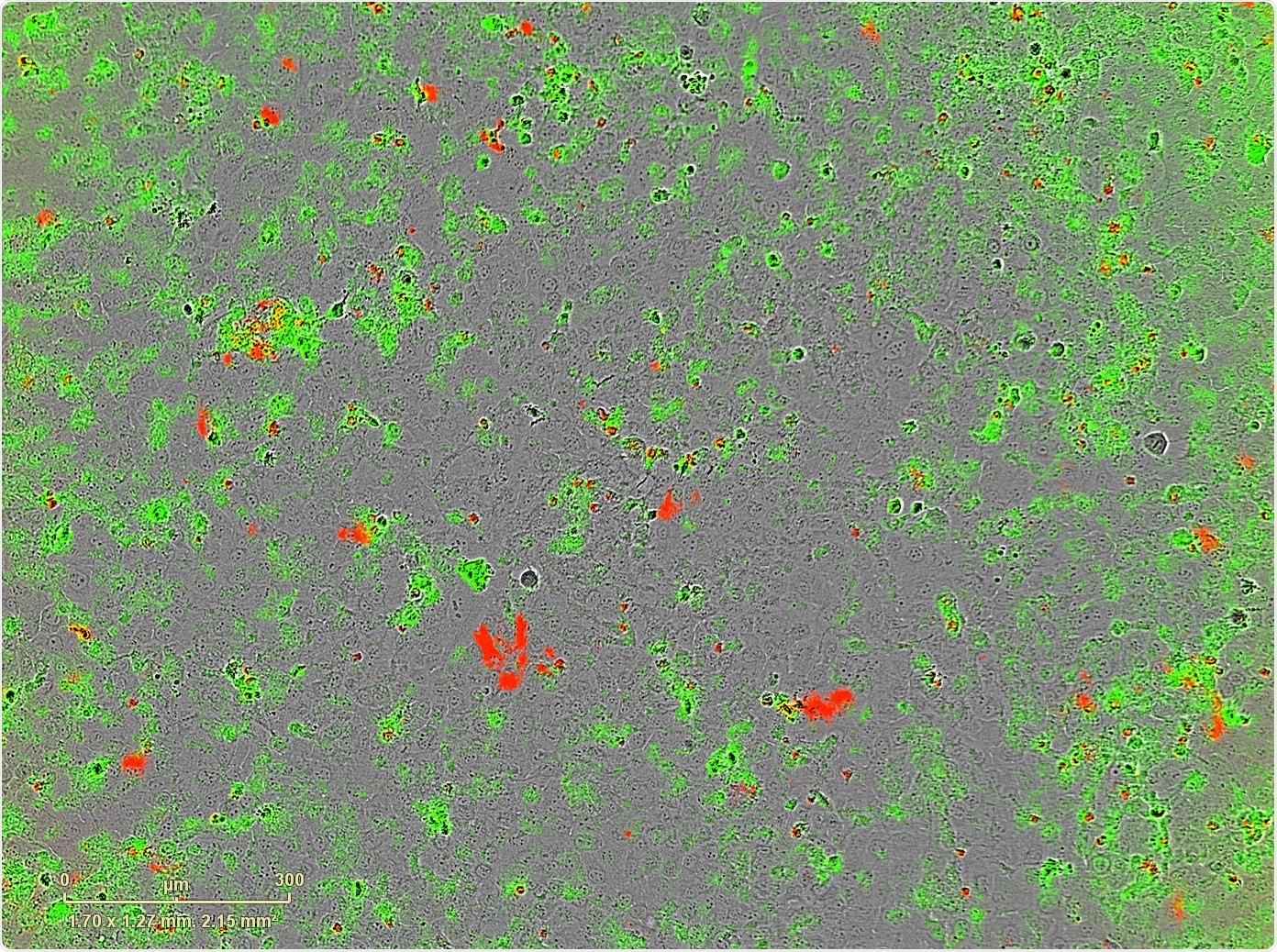
Now you don't have to do that, we can jump into the images four hours after plating to see how consistent we were, and if it was off, we'd just repeat it.
Another advantage that we weren’t able to achieve with our previous methods is the ability to investigate changes in cell morphology, especially when it comes to drugs, which can easily cause the cells to increase in size, to three, four times, which we can see now see quite plainly.
What would you like to see incorporated in the IncuCyte system in the future to help you further your research?
We were interested at the time in the IncuCyte® Scratch Wound Cell Migration and Invasion system for invasion assays, because traditional Boyden Chambers have some disadvantages including difficulties in quantitating research from that as well.
Unfortunately, we didn't have the funds, but it is something that, if we were able to get the funds in the future, I'd love to have.
What are the future aims for your research?
We're very interested in metastasis, in various different cancers. We also work on colon cancer, to a lesser extent, where we have some candidates for invasion and metastasis drivers; we would be very interested in testing these in the IncuCyte system.
Where can readers find more information?
- https://www.mdanderson.org/research/departments-labs-institutes/labs/kwong-laboratory.html
- https://www.essenbioscience.com/en/products/incucyte/
About Prof Lawrence Kwong
- 2007 first targeted rat model of colon cancer (PNAS)
- 2012 identification of MEK+CDK4/6 combination for NRAS melanoma (Nat Med), leading to clinical trial
- 2015 identification of BRAF inhibitor resistance protein patterns in human samples (JCI)
- 2017 whole exome sequencing analysis of mouse melanoma models and BRAF inhibitor resistance (Cell Reports)
- 2017 co-chair of TCGA cholangiocarcinoma analysis group (Cell Reports)
- 2017 co-corresponding author, metastatic colon cancer mouse model (Genes & Dev)
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.





















.png)












No hay comentarios:
Publicar un comentario