BMC Biochemistry
Structural similarities and functional differences clarify evolutionary relationships between tRNA healing enzymes and the myelin enzyme CNPase
BMC BiochemistryBMC series – open, inclusive and trusted201718:7
© The Author(s). 2017
- Received: 19 March 2017
- Accepted: 10 May 2017
- Published: 16 May 2017
Abstract
Background
Eukaryotic tRNA splicing is an essential process in the transformation of a primary tRNA transcript into a mature functional tRNA molecule. 5′-phosphate ligation involves two steps: a healing reaction catalyzed by polynucleotide kinase (PNK) in association with cyclic phosphodiesterase (CPDase), and a sealing reaction catalyzed by an RNA ligase. The enzymes that catalyze tRNA healing in yeast and higher eukaryotes are homologous to the members of the 2Hphosphoesterase superfamily, in particular to the vertebrate myelin enzyme 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase).
Results
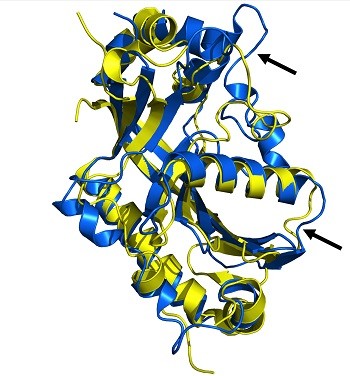
We employed different biophysical and biochemical methods to elucidate the overall structural and functional features of the tRNA healing enzymes yeast Trl1 PNK/CPDase and lancelet PNK/CPDase and compared them with vertebrate CNPase. The yeast and the lancelet enzymes have cyclic phosphodiesterase and polynucleotide kinase activity, while vertebrate CNPase lacks PNK activity. In addition, we also show that the healing enzymes are structurally similar to the vertebrate CNPase by applying synchrotron radiation circular dichroism spectroscopy and small-angle X-ray scattering.
Conclusions
We provide a structural analysis of the tRNA healing enzyme PNK and CPDase domains together. Our results support evolution of vertebrate CNPase from tRNA healing enzymes with a loss of function at its N-terminal PNK-like domain.
Keywords
- Cyclic phosphodiesterase
- Polynucleotide kinase
- Substrate specificity
- Protein structure
- 2H family
- Evolution
- tRNA splicing


































No hay comentarios:
Publicar un comentario