 Epigenetics & Chromatin
Epigenetics & ChromatinHigh-resolution mapping of centromeric protein association using APEX-chromatin fibers
- Received: 7 August 2018
- Accepted: 26 October 2018
- Published: 16 November 2018
Abstract
Background
The centromere is a specialized chromosomal locus that forms the basis for the assembly of a multi-protein complex, the kinetochore and ensures faithful chromosome segregation during every cell division. The repetitive nature of the underlying centromeric sequence represents a major obstacle for high-resolution mapping of protein binding using methods that rely on annotated genomes. Here, we present a novel microscopy-based approach called “APEX-chromatin fibers” for localizing protein binding over the repetitive centromeric sequences at kilobase resolution.
Results
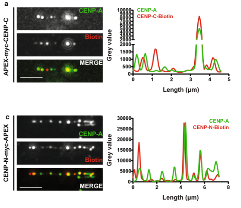
By fusing centromere factors of interest to ascorbate peroxidase, we were able to label their binding profiles on extended chromatin fibers with biotin marks. We applied APEX-chromatin fibers to at least one member of each CCAN complex, most of which show a localization pattern different from CENP-A but within the CENP-A delineated centromeric domain. Interestingly, we describe here a novel characteristic of CENP-I and CENP-B that display extended localization beyond the CENP-A boundaries.
Conclusions
Our approach was successfully applied for mapping protein association over centromeric chromatin, revealing previously undescribed localization patterns. In this study, we focused on centromeric factors, but we believe that this approach could be useful for mapping protein binding patterns in other repetitive regions.
Keywords
- Centromere
- Chromatin fibers
- CENP-A
- CCAN
- Kinetochore
- APEX


































No hay comentarios:
Publicar un comentario