Antibiotic Resistance Lab Network
Beginning in fall 2016, CDC’s Antibiotic Resistance Laboratory Network (AR Lab Network) will provide the infrastructure and lab capacity for seven regional labs to detect and support response to resistant organisms recovered from human samples. This network will complement additional investments to combat antibiotic resistance for labs in all states and five major cities/territories. These collective investments are a part of CDC’s Antibiotic Resistance Solutions Initiative, an expansion of CDC’s efforts to combat antibiotic resistance and implement the National Action Plan. This ambitious approach transforms much of the current laboratory landscape by closing the gap between hospital capabilities and data needed to combat AR.
As part of these investments, state health department labs designated as regional labs will be able to detect existing and emerging types of antibiotic resistance with gold-standard lab capacity, investigate emerging resistance faster and more effectively, and generate stronger data for improved infection control among patients to prevent and combat future resistance threats.
How the AR Lab Network Works
When new resistance threats or outbreaks are detected within healthcare facilities or state and local labs, regional labs will provide support, where needed, to characterize, support response, and track these discoveries. Since outbreak response varies by state, the support launched by the AR Lab Network may also vary by state or threat discovered. The regional labs will ensure more consistent and improved communication, coordination, and tracking at all levels—across healthcare facilities, state health departments, and CDC—every time.
The AR regional labs will work together with CDC and state and local health department labs to:
- Detect new resistance and provide better big-picture trend tracking to create pathogen-specific solutions and support national public health strategies
- Inform outbreak response when AR threats, like CRE, are reported, working together with state and local labs
- Prevent and combat future AR threats by creating better data
- Support innovations in antibiotic and diagnostic development. Samples from the labs will be made available through the FDA-CDC AR Isolate Bank, which researchers can use to develop earlier diagnoses and more effective treatment options.
The AR Lab Network regional labs will also conduct special threat assessments, as requested, for urgent, serious, and concerning threats identified by CDC, like Clostridium difficile [PDF - 5.24 MB], Methicillin-resistant Staphylococcus aureus [PDF - 5.24 MB] (MRSA), Vancomycin-resistant [PDF - 5.24 MB] Staphylococcus aureus (VRSA), and vancomycin-resistant enterococci [PDF - 5.24 MB] (VRE).
About the AR Lab Network Regional Labs
The AR regional labs have been strategically placed in regions of the United States to help track changes in resistance and help identify outbreaks with local technical support. Locally, the 2016 AR regional lab efforts will be coordinated by state health departments in Maryland, Minnesota, New York, Tennessee, Texas, Washington, and Wisconsin. Regions are defined by an existing regionalization scheme used by PulseNet.
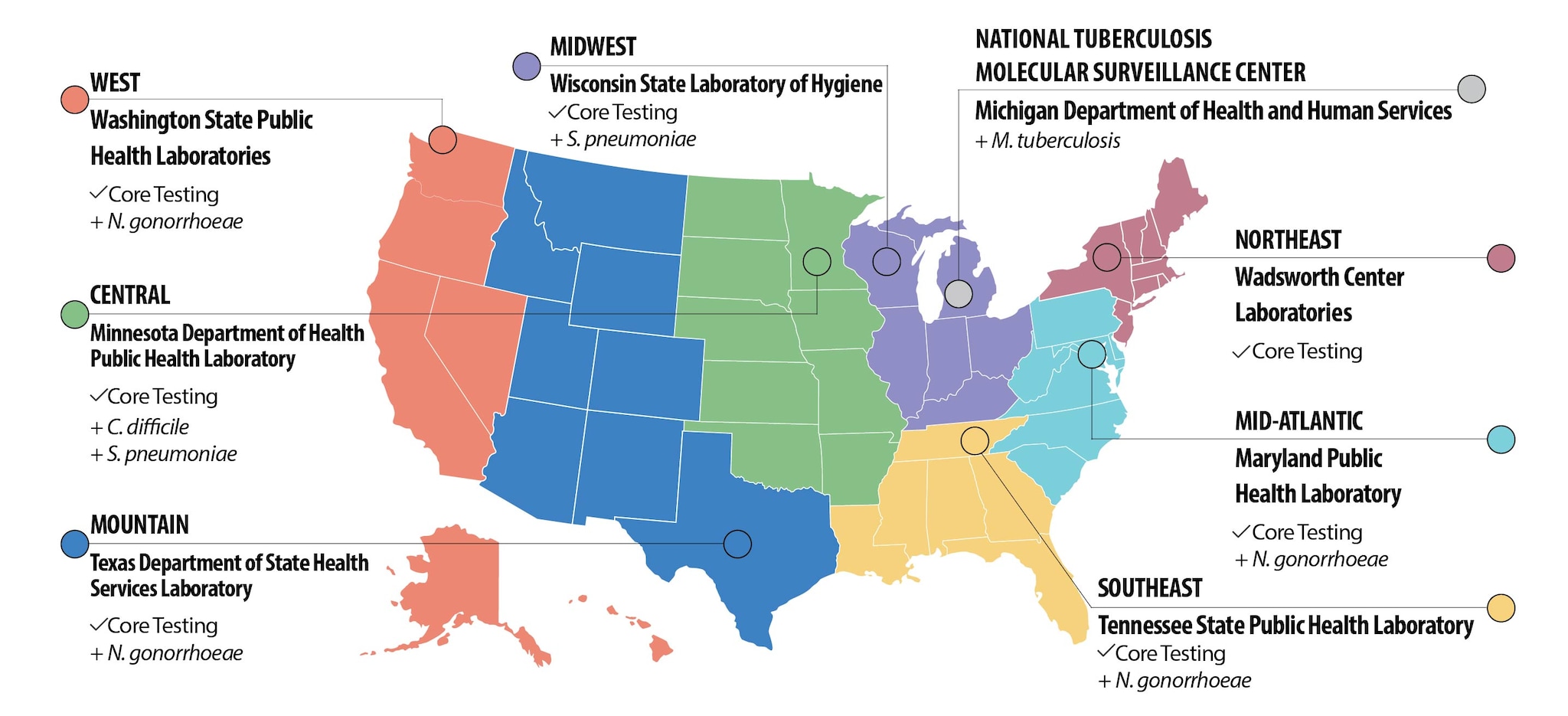
All regional labs will perform core testing for their region, including:
- Molecular testing to detect colonization of carbapenem-resistant Enterobacteriaceae (CRE)
- Threat assessments, special threat assessments by request on new or known threats like MRSA, VRE, and VRSA
- Isolate collection for use in CDC’s AR Isolate Bank and whole genome sequencing projects
Select labs will provide additional testing to support nationwide needs, including:
- Fungal susceptibility of Candida species to identify emerging resistance
- C. difficile special projects
- Increased testing of Neisseria gonorrhoeae for antimicrobial susceptibility
- Reflex Culture pilot with Salmonella and Entertoxigenic E. coli
- Antimicrobial susceptibility and serotyping of multidrug-resistant Streptococcus pneumoniae
In addition to this regional lab testing capacity, all 50 states and five cities will receive investments as part of CDC’s AR Solutions Initiative to increase capabilities to test for carbapenem-resistant Enterobacteriaceae (“nightmare bacteria”) and perform whole genome sequencing on all Salmonella isolates. These advancements will allow for faster identification and response to outbreaks. The regional labs will complement these activities done locally by providing additional or confirmation testing and outbreak support for these pathogens, when needed.
For more information about the AR Lab Network, email ARLN@cdc.gov.
AR Lab Network Factsheet
Infographic Details
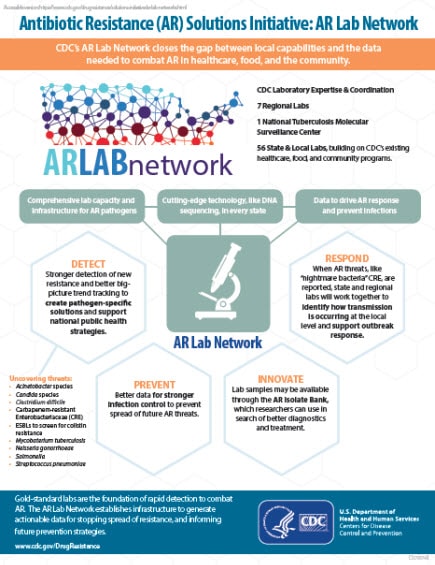
Antibiotic Resistance (AR) Solutions Initiative: AR Lab Network
CDC’s AR Lab Network closes the gap between hospital capabilities and data needed to combat AR−a network of state and regional labs fully equipped to detect resistance.
CDC’s AR Lab Network:
- Comprehensive lab capacity for antibiotic-resistant pathogens
- Gold standard labs and cutting-edge technology at local level
- Faster outbreak detection and response, better tracking of resistance
- Real-time, actionable data to prevent and combat future AR threats
Detect
Stronger detection of new resistance and better big-picture trend tracking to create pathogen-specific solutions and support national public health strategies
Respond
When AR threats, like “nightmare bacteria” or CRE, are reported, state and regional labs will work together to identify how transmission is occurring at the local level and support outbreak response
Prevent
Better data for stronger infection control to prevent and combat future AR threats
Innovate
Lab samples will be made available through the AR Isolate Bank, which researchers can use to develop earlier diagnoses and more effective treatment options
Using AR Isolate Bank samples:
- Pharmaceutical companies can test and create new antibiotics
- Biotech and diagnostics can design next generation clinical tests
- Researchers can study emerging resistance and investigate spread






















.jpg)












No hay comentarios:
Publicar un comentario