AMD Projects: Attacking Legionnaires’ Disease
Rapid molecular detection of Legionella for outbreak response
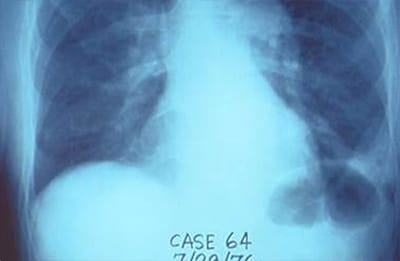
First discovered in 1976 at an American Legion convention in Philadelphia, Pennsylvania, Legionnaires’ disease continues to cause outbreaks in the United States. Even though it is a leading cause of potable waterborne outbreaks in the United States, reliable, rapid, and consistent analysis techniques have not been developed.
Available genetic techniques to determine the most likely source of transmission require matching clinical and environmental samples and can be used for only 1 of the more than 60 species of Legionella. Right now, it can take more than 3 weeks to completely identify the bacterium, if a specimen is even collected from a patient. These hurdles leave many cases and outbreaks uncharacterized. State and local health departments often rely on CDC laboratories because they do not have the specialized skills and extensive resources required to analyze Legionella specimens.
CDC is creating a database of Legionella genomes that will improve capacity for identifying and comparing outbreak-causing strains.
Advanced molecular detection (AMD) techniques are revolutionizing how assessments are made about the source of disease transmission, which is the cornerstone of controlling Legionella outbreaks. Rapid identification of a source will allow faster implementation of prevention efforts. This project will help streamline methods for state and local public health laboratories in investigating their own outbreaks leading to a quicker public health response. Ultimately, the methods developed through this project are the first critical steps in being able to detect Legionella straight from environmental or clinical samples, reducing laboratory and outbreak response time.
2017 Project Update
Since the project began, investigators used AMD to develop and implement a new method called whole genome Multilocus Sequence Typing (wgMLST) to quickly compare Legionella pneumophila strains. Previously researchers could only characterize small areas of DNA at once. Using wgMLST, researchers are currently able to look at several thousand specific sections (alleles) of DNA. The data from wgMLST can be shared easily between public health laboratories investigating an outbreak allowing for reliable, rapid, and consistent analysis. This new testing method, along with another type of test—single nucleotide polymorphism (SNP)—that looks at variations in shared DNA regions, were evaluated through a joint project with the New York State Department of Health. Both wgMLST and SNP analysis allow for a better view of DNA than traditional methods helping researchers identify new differences between strains. Additional laboratories will test these new methods in the upcoming year. Researchers will also continue to expand the wgMLST database by adding hundreds of new informative alleles as more highly diverse strains of Legionella are sequenced. With these improved and standardized methods of analyzing isolates, laboratories will be able to match environmental samples and clinical specimens quickly and accurately. This will allow state and local public health laboratories to share results faster and help prevent additional cases of Legionnaires’ disease.
PRINCIPAL INVESTIGATOR PROFILE
Jonas M. Winchell, PhD
Chief, Pneumonia Response and Surveillance Laboratory
Respiratory Diseases Branch
Division of Bacterial Diseases
National Center for Immunization and Respiratory Diseases
Respiratory Diseases Branch
Division of Bacterial Diseases
National Center for Immunization and Respiratory Diseases
Jonas M. Winchell, PhD is the chief of the Pneumonia Response and Surveillance Laboratory within the Division of Bacterial Diseases’ Respiratory Diseases Branch in CDC’s National Center for Immunization and Respiratory Diseases.
Winchell serves as an international and domestic consultant and technical expert on respiratory disease outbreaks that involve pathogens like Legionella and Mycoplasma, among others. He provides technical and laboratory support to determine which pathogen is causing an outbreak, including some that are novel or rare.
Winchell is leading CDC’s efforts to develop new ways to quickly identify these pathogens. Using sequence-based tests, his team of scientists will be able to detect a wide variety of respiratory pathogens, along with their important characteristics such as strain type and antibiotic resistance patterns. He is also overseeing efforts to create a whole genome pipeline for Legionella, which will improve laboratory capacity for identifying outbreak-causing strains of this deadly disease. Both of these projects will help CDC and public health officials better control respiratory disease outbreaks and protect the public’s health.
Winchell completed his Bachelor of Science in physiology and neurobiology, Master of Science in molecular and cell biology, and PhD in molecular and cell biology at the University of Connecticut. In 2013, he was recognized for his work in a Legionella outbreak investigation with receipt of an Excellence in Public Health Protection award from CDC’s National Center for Immunization and Respiratory Diseases. He has co-authored more than 50 scientific publications.


Last Posted: Mar 08, 2017
- Rapid molecular detection of Legionella for outbreak response
- Seeding and establishment of Legionella pneumophila in hospitals; implications for genomic investigations of nosocomial Legionnaires' disease.

David Sophia et al. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2017 Feb - A Large Community Outbreak of Legionnaires' Disease Associated With a Cooling Tower in New York City, 2015.

Weiss Don et al. Public health reports (Washington, D.C. : 1974) 2017 Jan 33354916689620 - Complete Genome Sequences of Legionella pneumophila subsp. fraseri Strains Detroit-1 and Dallas 1E.

Raphael Brian H et al. Genome announcements 2017 Feb 5(5) - Genomic Analysis Reveals Novel Diversity among the 1976 Philadelphia Legionnaires' Disease Outbreak Isolates and Additional ST36 Strains.

Mercante Jeffrey W et al. PloS one 11(9) e0164074 - Genomic Analysis Reveals Novel Diversity among the 1976 Philadelphia Legionnaires’ Disease Outbreak Isolates and Additional ST36 Strains

Mercante JW, et al. PLoS ONE, Sep 29, 2016. - Dynamics of genome change among Legionella species.

Joseph Sandeep J et al. Scientific reports 2016 633442 - Complete Genome Sequences of the Historical Legionella pneumophila Strains OLDA and Pontiac.

Mercante Jeffrey W et al. Genome announcements 2016 4(4) - Legionella DNA Markers in Tap Water Coincident with a Spike in Legionnaires’ Disease in Flint, MI

Schwake DO, et al. Environ Sci Technol Lett, Jul 8, 2016. - Confirming Legionnaires’ disease outbreak by genome-based method, Germany, 2012. Emerg Infect Dis. 2016 Jul
Burckhardt F, et al. Emerg Infect Dis, Jul 2016. - Rapid investigation of cases and clusters of Legionnaires' Disease in England and Wales using direct molecular typing.

Mentasti Massimo et al. Journal of medical microbiology 2016 Apr - Heated birthing pools as a source of Legionnaires' disease.

Collins S L et al. Epidemiology and infection 2016 Mar (4) 796-802 - Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires' disease incidents.

Moran-Gilad J et al. Euro Surveill. 2015 (28) - Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires' disease.

Gomez-Valero Laura et al. Genome Biol. 2014 (11) 505 - Gene flow in environmental Legionella pneumophila leads to genetic and pathogenic heterogeneity within a Legionnaires' disease outbreak.

McAdam Paul R et al. Genome Biol. 2014 (11) 504 - Genomic characterization of a large outbreak of Legionella pneumophila serogroup 1 strains in Quebec City, 2012.

Lévesque Simon et al. PLoS ONE 2014 (8) e103852






















.png)












No hay comentarios:
Publicar un comentario