
New tool records and tracks microbiome growth
Over the past years, the human microbiome has gained immense popularity due to its role in shaping one’s health. It is essential for human development, nutrition, and immunity. That’s why many studies have focused on how to improve one’s health by targeting or enhancing the microbiome.
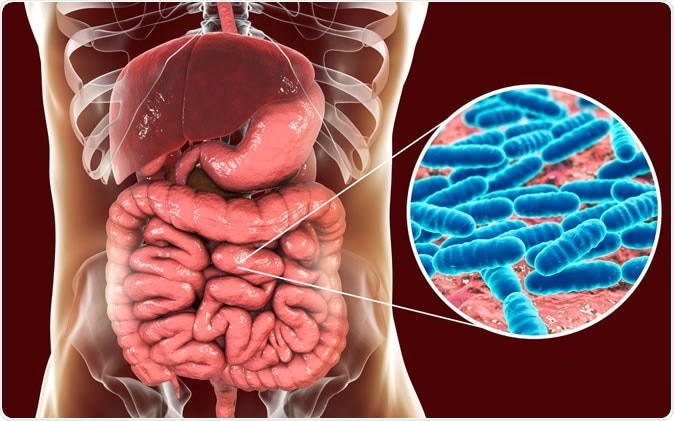
Normal flora of small intestine - Illustration Credit: Kateryna Kon / Shutterstock
The bacteria living in humans are not invaders but beneficial colonizers. They provide a wide array of health benefits. Any alteration in the balance of these species has been tied to autoimmune diseases like rheumatoid arthritis, multiple sclerosis, diabetes, fibromyalgia, and muscular dystrophy.
Many studies have dealt with the microbiome, but one part of its study that makes it hard for observation is how the flora changes over time in response to different stimuli. Currently, most scientists study the microbiome by extracting the bacteria from fecal samples. From there, they sequenced the genomes. But, one of the limitations of this type of experiment is that the vital information about the changes in the microbiome occurring in the gut is lost. Scientists, therefore, won’t have a complete picture of the dynamics of the human flora.
A team of researchers at the Wyss Institute for Biologically Inspired Engineering at Harvard University and Harvard Medical School (HMS) has found a way to solve the problem, as they were able to formulate bacterial genes that have been designed to determine and record the changes happening in the bacterial populations in the gut. They first tried the tool on mice models in the laboratory with single-cell precision. This new tool can pave the way for designing a more complex and comprehensive tool to be used for diagnostics and in formulating novel therapeutics.
Microbiome clock
Published in the journal Nature Communications, the study shows how the new algorithm can detect and track the changes happening in the gut flora populations. The system uses an oscillating gene circuit, dubbed as a repressilator, which is a type of genetic clock used to measure and gauge bacterial growth.
It contains three bacterial genes used to code for three proteins, namely the Tet Repressor proteins, lacl, and cl. These proteins block the expression of one of the other proteins. In the cycle, the genes are tied or connected into a negative feedback loop; wherein when one of the repressor proteins concentrations fall below normal, the repressed protein will be expressed. Hence, it blocks the expression of the third protein, and the cycle goes on.
So, when the scientists introduce the genes into the bacteria, the negative feedback loop cycle numbers that are completed can become a record of mitosis rate or how many cell divisions the bacteria have completed, more like a clock or timer.
“Imagine if you had two people wearing two different watches, and the second hand on one person's watch was moving twice as fast as the other person. If you stopped both watches after one hour, they wouldn't agree on what time it was, because their measurement of time varies based on the rate of the second hand's movement. In contrast, our repressilator is like a watch that always moves at the same speed, so no matter how many different people are wearing one, they will all give a consistent measurement of time. This quality allows us to study the behavior of bacteria in the gut more precisely,” David Riglar from the Wyss Institute and the Imperial College London, said.
“Imagine if you had two people wearing two different watches, and the second hand on one person's watch was moving twice as fast as the other person. If you stopped both watches after one hour, they wouldn't agree on what time it was, because their measurement of time varies based on the rate of the second hand's movement. In contrast, our repressilator is like a watch that always moves at the same speed, so no matter how many different people are wearing one, they will all give a consistent measurement of time. This quality allows us to study the behavior of bacteria in the gut more precisely,” David Riglar from the Wyss Institute and the Imperial College London, said.
To create the timer, the researchers partnered each of the three repressor proteins to a fluorescent molecule. They made the RINGS (Repressilator-based Inference of Growth at Single-cell level), which is an imaging workflow that can track or record the specific protein that is expressed at various time points throughout the growth of the bacteria.
With the use of RINGS, the researchers were able to successfully record the number of mitoses or cell divisions in many bacterial species. The new tool has helped solve a particular problem and at the same time, provide a platform that can help study the gut microbiome more. Also, it can open the door to new therapeutics and new diagnostic tests that may help curb various diseases associated with the gut microbiome imbalance or dysbiosis.
Journal reference:






















.png)












No hay comentarios:
Publicar un comentario