
Volume 23, Number 5—May 2017
Dispatch
Regional Transmission of Salmonella Paratyphi A, China, 1998–2012
On This Page
Abstract
To explore transmission patterns and genetic relationships of Salmonella enterica serovar Paratyphi A in China, we conducted a genome-wide single-nucleotide polymorphism analysis on the strains in the 4 provinces in which incidence was highest during 1998–2012. Markedly phylogeographic clustering suggested regional virus circulation after introduction from areas in southeastern China.
In Asia, incidence of paratyphoid fever remains high (1). In the mid-1990s, the number of paratyphoid fever cases in Asia caused by Salmonella enterica serovar Paratyphi A started to increase (2–4). In 2000, an estimated 5.41 million cases occurred; areas where incidence was highest (i.e., >100 cases/100,000 population per year) included south-central and Southeast Asia (5). Since 1998, the incidence of paratyphoid fever in Asia and the world has been highest in China, ranging from 0.08 to 192.5 cases/100,000 population annually (6); the provinces in which incidence is highest are Guangxi, Guizhou, Yunnan, and Zhejiang (7).
Information about the transmission routes and risk factors for infection could be used to improve the control strategies and measures for paratyphoid fever. Laboratory-based pathogen molecular subtyping, particularly genome-wide single-nucleotide polymorphism (SNP) analysis, can markedly improve outbreak detection, source tracing, and understanding of the epidemic modes. In this study, we analyzed genome-wide SNP and epidemiologic data from Salmonella Paratyphi A strains isolated from the China provinces where incidence was highest over a long period (1998–2012) and detected region-limited clone expansion in the epidemic provinces.
In 1998, the incidence of typhoid/paratyphoid fever in China was 4.82 cases/100,000 population (60,146 cases reported); this measure has since decreased annually to 0.88/100,000 (11,890 cases) in 2012 (China Information System for Disease Control and Prevention, unpub. data). Typhoid/paratyphoid fever cases in Guizhou, Yunnan, Zhejiang, and Guangxi Provinces accounted for 45.8% (in 1998) to 76.5% (in 2001) of all cases in China (Technical Appendix[PDF - 314 KB - 6 pages] Figure).
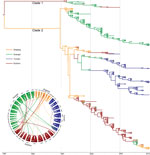
Figure 1. Phylogenetic tree of Salmonella enterica serovar Paratyphi A strains isolated from China, 1998–2012. The branches are colored according to inferred location. Inset: potential transmission of Salmonella Paratyphi A strains isolated from...
To analyze the genomic epidemiology of paratyphoid fever in these provinces, we first selected 96 Salmonella Paratyphi A strains circulating in 15 provinces in China during 1998–2011 (Technical Appendix[PDF - 314 KB - 6 pages] Table 1). Strains were isolated from hospitalized patients suspected of having typhoid/paratyphoid fever and were maintained in the strain bank of the Chinese Center for Disease Control and Prevention. We then conducted genome-wide SNP genotyping by using the iPLEX Gold assay (Sequenom Inc., San Diego, CA, USA) with 2,343 SNPs obtained from 7 genomes sequenced in a previous study (8) and 17 genomes of Salmonella Paratyphi A strains sequenced in this study. We obtained 112 phylogenetically informative SNPs (including 57 nonsynonymous SNPs) (Technical Appendix[PDF - 314 KB - 6 pages] Table 2), which were further analyzed in 335 Salmonella Paratyphi A strains (Technical Appendix[PDF - 314 KB - 6 pages]Table 1) isolated from the provinces where incidence was highest (i.e., Guangxi, Guizhou, Yunnan, and Zhejiang) during 1998–2012 by using the iPLEX Gold assay. The population history of Salmonella Paratyphi A was estimated by using BEAST version 2.1.3 (http://beast.bio.ed.ac.uk/), and the maximum clade credibility tree was summarized by using TreeAnnotator and visualized by using FigTree version 1.4.2 (both within BEAST). The consensus tree (Figure 1) showed that all strains fell into 2 main clades: clade 1 consisted of 16 strains isolated from Yunnan, Guizhou, and Guangxi Provinces during 1998–2007; clade 2 consisted of the strains that were most common and widespread in these 4 provinces during 1998–2012. In clade 2, at least 3 subclades were formed, which were markedly characterized by geographic clustering according to province (Figure 1), suggesting intraprovince transmission of the different clones. In addition, the earlier strains in the root of each major subbranch were isolated mainly from Zhejiang, and in the years before 2005, some strains from Guangxi were also mixed in the Guizhou branch.
On the basis of the trees, we further determined from/to transmission of Salmonella Paratyphi A by using Circos (9) (Figure 1). The same SNP genotypes of Salmonella Paratyphi A strains were preferentially transmitted within a single province from year to year, whereas the strains from Zhejiang were frequently transmitted to Guizhou and Guangxi, particularly during 1998–2002. The transmission between Guangxi and Guizhou was markedly more frequent before 2004 and decreased after 2005. After 2005, we found no transmission from Yunnan to other provinces.
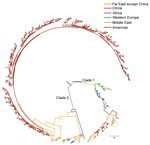
Figure 2. Phylogenetic tree of Salmonella enterica serovar Paratyphi A strains in China and worldwide. The branches are colored according to the inferred location. Scale bar indicates number of years.
We also extracted information for 112 SNPs from 127 genomes of the worldwide Salmonella Paratyphi A isolates in GenBank (10) and constructed a phylogenetic tree by combining these data with data from the 335 strains from China obtained in this study (Figure 2). The international strains fell into 2 clades, and the strains from Southeast and southern Asia were positioned much closer to the root of the strains from China, suggesting that the potential source of Salmonella Paratyphi A in China might be India or Indonesia.
The genome-wide SNP phylogeny provided more accurate insights into the variation of Salmonella Paratyphi A strains in China. In Guizhou, Guangxi, and Yunnan Provinces, which are geographically adjacent, Salmonella Paratyphi A has existed for many years. Although we had speculated that the organism might show a mixture of genetic patterns, the phylogenetic tree showed that epidemic strains from different provinces gradually accumulated their own mutations to evolve and form obvious geographic branches. In earlier years of the study period (1998–2002), the epidemic strains from Guangxi and Guizhou Provinces might have originated from early epidemic strains from Zhejiang Province. The level of economic development in Zhejiang Province is high, whereas in Guangxi, Guizhou, and Yunnan Provinces it is lower; the rural population from these 3 provinces migrates frequently to work in the economically developed southeastern coastal areas in China, including Zhejiang (11), Jiangsu, and Guangdong Provinces. According to the fifth national census conducted in 2000 (http://www.stats.gov.cn/tjsj/pcsj/rkpc/5rp/index.htm) and the sixth conducted in 2010 (http://www.stats.gov.cn/tjsj/pcsj/rkpc/6rp/indexch.htm), the migration data within the 4 provinces showed this population movement trend (Technical Appendix[PDF - 314 KB - 6 pages] Table 3). At irregular intervals, migrant workers, mainly those who are young and middle-aged, return to their hometown for family reunions.
In the mid-1990s, paratyphoid fever became an emerging problem in Zhejiang Province; during 1997–2005, incidence was 8.61 cases/100,000 population (12). In those years, managing ex situ healthcare and medical treatments in China was problematic. When migrant workers got ill, they seldom sought medical treatment at the hospital in the city in which they worked; rather, they bought medicine at a chemist’s shop or returned to their hometown for treatment (Zhang Q. The study on the health seeking behavior of migrant workers [master’s thesis]. China: Shaanxi Normal University; 2012).
Because of lack of medical treatment in hospitals, migrant workers who become infected with Salmonella Paratyphi A easily become chronic carriers. Therefore, Salmonella Paratyphi A might be transmitted to Guangxi, Guizhou, and Yunnan Provinces via a migrating infected population, including patients and carriers. In addition, these 3 provinces are mainly mountainous, and the population flow among these provinces is limited by their lower economic development and inaccessibility. Therefore, the transmission pattern in these regions could be closely associated with the southeastern coastal areas, where the level of economic development is higher, and transmission among these 3 provinces could be absent. Moreover, in these paratyphoid-epidemic provinces, most of the overall population lives in rural agricultural areas. Given the combination of poor water and food hygiene with a hot and humid climate, the epidemic clones of Salmonella Paratyphi A could persist for a long time after being introduced into these areas.
In summary, we identified the evolution and transmission mode of paratyphoid fever in the China provinces where incidence is highest. Populations migrating to southeastern China probably mediated the transmission of Salmonella Paratyphi A. Considering the obvious regional clone expansion in these provinces, the local natural, social, and economic conditions need to be investigated for their potential roles in the spread of paratyphoid fever and for the development of intervention strategies.
Dr. Lu is associate professor at the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing. Her research interests focus on molecular epidemiology, population genetics, and evolution of pathogenic enteric bacteria.
Acknowledgment
This work was supported by the National Key Basic Research Program (2015CB554201), the National Key Research and Development Program (2016YFC1200103) of the Ministry of Health of China and by the Science Foundation (2014SKLID203) of the State Key Laboratory of Infectious Disease Prevention and Control, China.
References
- Crump JA, Mintz ED. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50:241–6. DOIPubMed
- Dong BQ, Yang J, Wang XY, Gong J, von Seidlein L, Wang ML, et al. Trends and disease burden of enteric fever in Guangxi province, China, 1994-2004. Bull World Health Organ. 2010;88:689–96. DOIPubMed
- Karkey A, Thompson CN, Tran Vu Thieu N, Dongol S, Le Thi Phuong T, Voong Vinh P, et al. Differential epidemiology of Salmonella Typhi and Paratyphi A in Kathmandu, Nepal: a matched case control investigation in a highly endemic enteric fever setting. PLoS Negl Trop Dis. 2013;7:e2391. DOIPubMed
- Ochiai RL, Wang X, von Seidlein L, Yang J, Bhutta ZA, Bhattacharya SK, et al. Salmonella paratyphi A rates, Asia. Emerg Infect Dis. 2005;11:1764–6. DOIPubMed
- Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–53.https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15298225&dopt=Abstract
- Arndt MB, Mosites EM, Tian M, Forouzanfar MH, Mokhdad AH, Meller M, et al. Estimating the burden of paratyphoid a in Asia and Africa. PLoS Negl Trop Dis. 2014;8:e2925. DOIPubMed
- Meiying Y, Weili L, Wei L, Biao K. Epidemics of typhoid and paratyphoid fever from 1995 through 2004 in China. Dis Surveill. 2005;20:401–3.
- Liang W, Zhao Y, Chen C, Cui X, Yu J, Xiao J, et al. Pan-genomic analysis provides insights into the genomic variation and evolution of Salmonella Paratyphi A. PLoS One. 2012;7:e45346. DOIPubMed
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. DOIPubMed
- Zhou Z, McCann A, Weill FX, Blin C, Nair S, Wain J, et al. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci U S A. 2014;111:12199–204. DOIPubMed
- Hu F. Researches on China’s rural labor migration: a literature survey [in Chinese]. Zhejiang Social Sciences. 2007;1:207–12.
- Qin S, Chen E, Xie S. Analysis on epidemiological characteristics of typhoid and paratyphoid fever from 1997 to 2006 in Zhejiang Province. Zhejiang Journal of Preventive Medicine. 2008;20:1–5.






















.png)











No hay comentarios:
Publicar un comentario