
Volume 25, Number 11—November 2019
Research
Serosurvey for Influenza D Virus Exposure in Cattle, United States, 2014–2015
Tables
Downloads
Article Metrics
Simone Silveira, Shollie M. Falkenberg , Bryan S. Kaplan, Beate Crossley, Julia F. Ridpath, Fernando B. Bauermann, Charles P. Fossler, David A. Dargatz, Rohana P. Dassanayake, Amy L. Vincent, Cláudio W. Canal, and John D. Neill
, Bryan S. Kaplan, Beate Crossley, Julia F. Ridpath, Fernando B. Bauermann, Charles P. Fossler, David A. Dargatz, Rohana P. Dassanayake, Amy L. Vincent, Cláudio W. Canal, and John D. Neill
Abstract
Influenza D virus has been detected predominantly in cattle from several countries. In the United States, regional and state seropositive rates for influenza D have previously been reported, but little information exists to evaluate national seroprevalence. We performed a serosurveillance study with 1,992 bovine serum samples collected across the country in 2014 and 2015. We found a high overall seropositive rate of 77.5% nationally; regional rates varied from 47.7% to 84.6%. Samples from the Upper Midwest and Mountain West regions showed the highest seropositive rates. In addition, seropositive samples were found in 41 of the 42 states from which cattle originated, demonstrating that influenza D virus circulated widely in cattle during this period. The distribution of influenza D virus in cattle from the United States highlights the need for greater understanding about pathogenesis, epidemiology, and the implications for animal health.
Influenza D virus (IDV; genus Deltainfluenzavirus, family Orthomyxoviridae) is an enveloped, single-stranded, negative sense RNA virus with 7 genome segments and 1 surface glycoprotein, the hemagglutinin-esterase fusion (HEF) protein (1,2). The first detection of IDV dates back to Oklahoma, USA, in 2011 from pigs exhibiting influenza-like disease (3), although retrospective seroprevalence data suggest the presence of IDV in goats in the United States before 2002 (4). Subsequently, IDV has been identified in low frequency in pigs in Italy (5,6) and Luxembourg (7). In addition, evidence suggests IDV circulates in other hosts such as small ruminants, camels, and buffalo in Togo, Kenya, and China (8,9) and small ruminants, feral swine, and equids in the United States (4,10,11).
Although IDV has been detected in other species, cattle appear to be the main reservoir (1,12). A variety of sample types and methods of detection have been used to determine the prevalence of IDV in different regions, in various ages, breeds, and numbers of cattle evaluated. The lack of consistency between the methods and cattle evaluated may be a contributing factor to variability in prevalence of IDV in different regions. Seroprevalence data have been reported in cattle from Luxembourg (7), Japan (13,14), the United States (1,15,16), Togo, Benin, and Morocco (9); the highest reported seropositive rate (80.2%) was in the United States (16) and Luxembourg (7) and the lowest (1.9%) in Benin (9). Serologic testing provides an indication of IDV exposure but is not a measure of active infections. IDV RNA from respiratory samples of cattle has been detected in several countries: the United States (1,15,17,18), Italy (5), France (19), Ireland (20), China (8,21), Japan (22), and Mexico (18). Studies from Mexico (18) reported the highest frequency of positive samples (29.7%) and China the lowest (0.7%) (21).
In both experimental and field infections with IDV, mild to moderate respiratory disease has been reported (23,24). In addition, IDV-positive samples are reported not only from cattle manifesting clinical signs associated with bovine respiratory disease but also from cattle that are asymptomatic and appear to be healthy (20–22). Experimental infection of calves demonstrated that IDV caused mild to moderate respiratory disease and that peak viral shedding occurred at 4–6 days postinfection; seroconversion was detected as early as day 6 postinfection (12,23,24). Whereas IDV infection by itself has been associated mainly with mild respiratory illness, IDV has also been implicated as a contributor to bovine respiratory disease complex (BRDC), which is the most costly disease affecting the US cattle industry (17,18,23,25).
Because there are no commercially available vaccines against IDV, positive serologic assays reflect natural exposure. Given the potential of IDV to contribute to BRDC, inclusion of IDV in vaccination programs has been debated. The frequency of IDV RNA–positive samples from US cattle is 4.8%–18% (1,15,17,18), and positive samples have been reported in the US cattle population since 2003 (16). The seropositive rate has been reported at 13.5%–80.2% (15,16); the Upper Midwest region has the highest seroprevalence. The wide variation of seroprevalence could be caused by differences in the age of the cattle evaluated or by differences across regions because of limited sample size and the focus on the Midwest and South Central regions of the country. We conducted a national serosurvey of cattle of a similar age to fully evaluate the potential role of IDV in BRDC infections and the effect of IDV on animal health and productivity.
Samples
We assessed 1,992 banked bovine serum samples for IDV-specific antibodies. The samples, collected between August 2014 and December 2015 as part of the US brucellosis surveillance program, were previously used to screen for ruminant pestivirus and bovine leukemia virus (BLV) exposure (26,27). We aimed to determine the seropositivity rate for IDV and retrospectively compare that rate with seropositivity rates for ruminant pestivirus and BLV from the same samples to identify regional patterns or differences in the US cattle population.
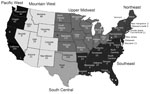
The serum samples came from both male and female cattle >2 years of age, raised in 42 states, and were randomly collected from 5 slaughter plants. The states were categorized into 6 regions as previously defined (26): Pacific West (PW), Mountain West (MW), Upper Midwest (UMW), South Central (SC), Northeast (NE), and Southeast (SE) (Figure 1). The number of samples taken in each slaughter plant, listed by state (California, Florida, Nebraska, Pennsylvania, Minnesota), was proportional to the total annual number of cattle >2 years of age that had been processed in that plant. All samples were previously reported as negative for brucellosis.
Virus Selection and Propagation
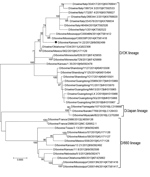
To select the IDV strain used for the hemagglutination inhibition (HI) assay, we performed phylogenetic analysis on HEF genes with IDV strains that circulated in the United States during the same period in which the samples used for this study were collected (Figure 2). We downloaded full-length HEF gene segment sequences (n = 39) from the Influenza Research Database () on September 28, 2018. We aligned full-length segments using the MAFFT plug-in for Geneious version 9.1.4 (Biomatters Ltd., ) with subsequent manual correction. We constructed a maximum-likelihood tree inferred in IQ-tree () using a general time-reversible model of nucleotide substitution combined with a gamma-distributed rate variation with statistical support generated through ultrafast bootstrap analysis (28,29). We chose a representative US strain, D/bovine/Kansas/14-22/12, showing a high amino acid similarity (96%–99.2%) with US strains detected during 2014–2015, and a high hemagglutination (HA) titer.
We maintained swine testicle cells (ATCC CRL-1746) used for propagation of IDV in MEM medium (Sigma Aldrich, ), supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (PAA Laboratories, Inc., ) and L-glutamine (ThermoFisher Scientific, ) antibiotic-antimycotic solution incubated at 37°C in a humid atmosphere of 5% CO2. We propagated the D/bovine/Kansas/14-22/12 strain, diluted 1:1,000 in swine testicle cells cultured in serum-free medium in the presence of TPCK-trypsin (0.1 µg/mL) and 5% bovine serum albumin, and incubated at 37°C for up to 4 days.
Serology
We performed the HI assay for detection of D/bovine/Kansas/14-22/12–specific antibodies in accordance with the specifications in the World Health Organization manual on animal influenza A virus diagnosis and surveillance (30). We treated 1:3 serum samples with receptor-destroying enzyme (Denka Seiken UK, ) at 37°C for 18 hours, heat inactivated it at 56°C for 1 h, and diluted it 1:10 with phosphate-buffered saline. We conducted the assay in duplicate, at room temperature and in V-bottom 96-well plates, starting at 1:10 and doing 2-fold serial dilutions to reach a 1:1,280 dilution. We added the serially diluted samples to the virus (8 hemagglutination units/50 µL) for 1 h, then incubated with 0.5% turkey red blood cells for 30 min. The endpoint titer was the reciprocal of the highest dilution of serum that demonstrated partial to full inhibition of hemagglutination. We determined that a serum with an HI titer >40 was seropositive according to previous IDV serosurveillance studies (4,15). We used a negative control (PBS), as well as a positive control consisting of a rabbit polyclonal antiserum generated against D/swine/OK/1334/2011, in the HI assay (1). To exclude the possible presence of nonspecific antibodies, we also performed HI with serum samples from 10 colostrum-deprived calves; all showed titers of 0, which confirmed negativity.
Statistical Analysis
We used GraphPad Prism 7 software (GraphPad Software, LLC, ) to statistically compare seropositive rates of IDV infection by χ2 test and geometric mean titers (GMT) by the Kruskal-Wallis and Mann-Whitney tests. We considered p<0.05 significant.
Of the 1,992 cattle serum samples tested by HI assay for detection of IDV-specific antibodies, 1,545 (77.5%) samples were positive; the overall GMT of positive samples was 230 (titers ranged from 40 to 1,280). We identified positive serum in samples from 41 of the 42 states tested (Table). The seropositivity rate was 25%–93.8% among the states and average GMT was 80–460. However, sample size was small in some of the states with low positivity, low titer, or both, which might have caused bias in the regional distribution.
We categorized the results by geographic region to compare the differences of seropositive rate and GMT. The seropositive rate range was 47.7%–84.6% (p<0.05) and GMT 110–260 (p<0.05) among the regions (Table). Mountain West region had the highest seropositive rate (84.6%) and GMT (260); Northeast region had the lowest seropositive rate (47.7%) and GMT (110).
Although IDV was described in pigs earlier than in cattle in the United States, subsequent reports of retrospective samples suggested that cattle are the natural reservoir (1,12). Because seroprevalence surveillance in US cattle had been conducted only at state or regional levels, we undertook a nationwide serologic survey to detect IDV antibodies in cattle. Our results clearly demonstrate that IDV circulated with high frequency in cattle in all regions of the United States during 2014–2015.
We observed regional variation in seropositive rate and GMT, although all regions had relatively high frequency. Overall, the Upper Midwest and Mountain West regions showed the highest seropositive rates and the highest antibody titers, and also encompassed the states with the highest GMT. A similar result was obtained in a pestivirus serologic study performed with the same serum samples; here too, the Mountain West region showed the highest number of antibody-positive animals and higher titers (26). Although it is not possible to establish the cause, both pestivirus and IDV serology follow a similar trend. Potential causes include herd size, which can exceed 1,000 animals in these areas, and the potential for livestock and wildlife species to commingle and facilitate virus transmission. Evidence indicates IDV can infect nonbovine hosts, such as sheep, goats, pigs, and equids, in the United States (4,10,31). However, the full range of susceptible hosts for IDV is unknown, and interspecies transmission has not been demonstrated among the known hosts.
Seroprevalence of IDV in small ruminants was reported in samples collected from the Mountain West and Upper Midwest regions, whereas samples from other regions were negative (4). Moreover, in the Upper Midwest region, a high percentage of small ruminants with high titers was described, and the farms where they were located were in close proximity to cattle farms (4). This issue needs to be explored further to understand the importance of IDV as a threat for animal health and whether this is an underlying factor for the increased seroprevalence of viral pathogens in regions that have greater potential for interspecies transmission.
In general, we observed lower titers and a lower percentage of positive animals in the Northeast and Southeast regions. These results are similar to those reported from the pestivirus serosurvey that also found these 2 regions to have the lowest titers and lowest number of cattle seropositive for BVDV (26). On the other hand, in the BLV serosurvey, the Northeast had the highest seropositive rate for BLV and the Mountain West the lowest seropositive rate (27). Although seroprevalence differences existed between BLV and the other viruses evaluated (pestivirus and IDV), these differences could be caused by limited number of samples collected in these regions, differences in the epidemiology of these viruses, or differences in herd management practices across the regions. Previous data of IDV exposure in cattle of different ages in Mississippi (Southeast region) reported a high seroprevalence in cattle >1 year of age (15). Discrepancies between the current study and the previous reports could be explained by the number of samples evaluated in each of these studies; only 4 samples originated from Mississippi in our study, whereas >500 cattle were sampled in a previous study (15). Although our study encompassed the entire United States, the limited number of samples from several states, and subsequently the regions they represent, may have caused underestimation or overestimation of the seropositive rate of IDV. Despite the limitations of our study, data indicate that IDV is widespread at rates similar to the regional or state data previously reported (15,16).
Our findings, combined with those from previous serosurveillance studies (15,16), confirm a high nationwide seroprevalence of IDV in US cattle populations. Because of the potential association of IDV with BRDC (17,18,23,31) and the dearth of vaccines to prevent IDV infection (12,32), concerns have been raised regarding the negative effect of IDV on animal health. A possible explanation for the high seropositive rate is that IDV is common in the respiratory tract of cattle; times of stress, immune attack, or environmental changes that affect the respiratory tract can increase viral shedding but might not cause disease. Unpublished diagnostic data from our laboratory show that IDV is detected more frequently in samples that are also positive for other respiratory pathogens than in those positive for IDV alone. This finding indicates that IDV can either predispose the respiratory tract or act as an opportunistic pathogen in concert with other pathogens to cause BRD. Further research, including co-infection studies, is needed to elucidate the full range of susceptible hosts and the dynamics of interspecies transmission to understand the contribution of IDV to BRDC. In summary, our serosurveillance study of bovine serum samples from 2014–2015 showed a high seropositivity rate for IDV in the United States; 41 of the 42 states from which cattle originated had seropositive animals. No IDV vaccine exists. IDV infection has also been implicated in BRDC, the most costly disease affecting the US cattle industry. Therefore, our findings may indicate an ongoing risk to animal health.
Dr. Silveira is a postdoctoral researcher at the Universidade de Caxias do Sul (UCS), Brazil. Her primary research interests are molecular epidemiology and evolution of emerging influenza viruses and pestiviruses.
Acknowledgments
We thank the staff at the Kentucky Federal Brucellosis Laboratory, including Christopher Cracraft, Shanna Wilburn, April Shannon, Lisa Burke, Megan Zinner, Phyllis Bays, and Jeffery Gifford, for logistic support in identifying and shipping serum samples to the laboratory for testing. We thank Mark Camacho for his technical assistance in setting up the collaboration between the US Department of Agriculture, Agricultural Research Service (USDA/ARS), and the laboratory. We also thank the staff at the National Animal Disease Center, including Kathryn McMullen, Patricia Federico, and Renae Lesan. Finally, we thank Kathy Simmons and the National Cattlemen’s Beef Association for their interest in this project and their work to promote cattle health research projects at USDA/ARS.
B.S.K. is supported by an appointment to the USDA/ARS Research Participation Program, administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy and USDA (contract no. DE-AC05-06OR23100). S.S. and C.W.C. received support from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brasil (CAPES) (finance code 001) and Conselho Nacional de Pesquisa–Brasil (CNPq).
This study was conducted at a USDA research facility and all funding was provided through internal USDA research dollars. This project is an intramural project of the USDA/ARS National Animal Disease Center (5030-32000-117-00D). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
References
- Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, et al. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5:e00031–14.
- International Committee on Taxonomy of Viruses. Taxonomy. 2017 [cited 2019 Aug 28].
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:
e1003176 . - Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet Microbiol. 2015;180:281–5.
- Chiapponi C, Faccini S, De Mattia A, Baioni L, Barbieri I, Rosignoli C, et al. Detection of influenza D virus among swine and cattle, Italy. Emerg Infect Dis. 2016;22:352–4.
- Foni E, Chiapponi C, Baioni L, Zanni I, Merenda M, Rosignoli C, et al. Influenza D in Italy: towards a better understanding of an emerging viral infection in swine. Sci Rep. 2017;7:11660.
- Snoeck CJ, Oliva J, Pauly M, Losch S, Wildschutz F, Muller CP, et al. Influenza D virus circulation in cattle and swine, Luxembourg, 2012–2016. Emerg Infect Dis. 2018;24:1388–9.
- Zhai S-L, Zhang H, Chen S-N, Zhou X, Lin T, Liu R, et al. Influenza D virus in animal species in Guangdong Province, southern China. Emerg Infect Dis. 2017;23:1392–6.
- Salem E, Cook EAJ, Lbacha HA, Oliva J, Awoume F, Aplogan GL, et al. Serologic evidence for influenza C and D virus among ruminants and camelids, Africa, 1991–2015. Emerg Infect Dis. 2017;23:1556–9.
- Nedland H, Wollman J, Sreenivasan C, Quast M, Singrey A, Fawcett L, et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health. 2018;65:e148–54.
- Ferguson L, Luo K, Olivier AK, Cunningham FL, Blackmon S, Hanson-Dorr K, et al. Influenza D virus infection in feral swine populations, United States. Emerg Infect Dis. 2018;24:1020–8.
- Hause BM, Huntimer L, Falkenberg S, Henningson J, Lechtenberg K, Halbur T. An inactivated influenza D virus vaccine partially protects cattle from respiratory disease caused by homologous challenge. Vet Microbiol. 2017;199:47–53.
- Horimoto T, Hiono T, Mekata H, Odagiri T, Lei Z, Kobayashi T, et al. Nationwide distribution of bovine influenza D virus infection in Japan. PLoS One. 2016;11:
e0163828 . - Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, et al. Influenza D virus infection in herd of cattle, Japan. Emerg Infect Dis. 2016;22:1517–9.
- Ferguson L, Eckard L, Epperson WB, Long LP, Smith D, Huston C, et al. Influenza D virus infection in Mississippi beef cattle. Virology. 2015;486:28–34.
- Luo J, Ferguson L, Smith DR, Woolums AR, Epperson WB, Wan XF. Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003-2004. Virology. 2017;501:88–91.
- Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol. 2015;89:1036–42.
- Mitra N, Cernicchiaro N, Torres S, Li F, Hause BM. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J Gen Virol. 2016;97:1771–84.
- Ducatez MF, Pelletier C, Meyer G. Influenza D virus in cattle, France, 2011-2014. Emerg Infect Dis. 2015;21:368–71.
- Flynn O, Gallagher C, Mooney J, Irvine C, Ducatez M, Hause B, et al. Influenza D virus in cattle, Ireland. Emerg Infect Dis. 2018;24:389–91.
- Jiang W-M, Wang S-C, Peng C, Yu J-M, Zhuang Q-Y, Hou G-Y, et al. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes. 2014;49:493–6.
- Mekata H, Yamamoto M, Hamabe S, Tanaka H, Omatsu T, Mizutani T, et al. Molecular epidemiological survey and phylogenetic analysis of bovine influenza D virus in Japan. Transbound Emerg Dis. 2018;65:e355–60.
- Ferguson L, Olivier AK, Genova S, Epperson WB, Smith DR, Schneider L, et al. Pathogenesis of influenza D virus in cattle. J Virol. 2016;90:5636–42.
- Salem E, Hägglund S, Cassard H, Corre T, Näslund K, Foret C, et al. Pathogenesis, host innate immune response and aerosol transmission of Influenza D virus in cattle. J Virol. 2019; 93:JVI.01853-18.
- Ng TFF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol. 2015;89:5340–9.
- Bauermann FV, Ridpath JF, Dargatz DA. A serosurvey for ruminant pestivirus exposure conducted using cattle sera collected for brucellosis surveillance in the United States. J Vet Diagn Invest. 2017;29:76–82.
- Bauermann FV, Ridpath JF, Dargatz DA. Bovine leukemia virus seroprevalence among cattle presented for slaughter in the United States. J Vet Diagn Invest. 2017;29:704–6.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
- World Health Organization. Manual for the laboratory diagnosis and virological surveillance of influenza. 2011 [cited 2019 Aug 28].
- US Department of Agriculture. Feedlot 2011 part IV: health and health management on US feedlots with a capacity of 1,000 or more head. 2013 [cited 2019 Aug 28].
- Wan Y, Kang G, Sreenivasan C, Daharsh L, Zhang J, Fan W, et al. A DNA vaccine expressing consensus hemagglutinin-esterase fusion protein protected guinea pigs from infection by two lineages of influenza D virus. 2018. J Virol. 2018;92:e00110–8.
Figures
Table
Cite This ArticleOriginal Publication Date: 9/30/2019





















.png)












No hay comentarios:
Publicar un comentario