
Volume 24, Number 7—July 2018
Dispatch
Guiana Dolphin Unusual Mortality Event and Link to Cetacean Morbillivirus, Brazil
On This Page
Kátia R. Groch , Elitieri B. Santos-Neto, Josué Díaz-Delgado, Joana M.P. Ikeda, Rafael R. Carvalho, Raissa B. Oliveira, Emi B. Guari, Tatiana L. Bisi, Alexandre F. Azevedo, José Lailson-Brito, and José L. Catão-Dias
, Elitieri B. Santos-Neto, Josué Díaz-Delgado, Joana M.P. Ikeda, Rafael R. Carvalho, Raissa B. Oliveira, Emi B. Guari, Tatiana L. Bisi, Alexandre F. Azevedo, José Lailson-Brito, and José L. Catão-Dias
Abstract
During November–December 2017, a mass die-off of Guiana dolphins (Sotalia guianensis) began in Rio de Janeiro, Brazil. Molecular and pathologic investigations on 20 animals indicated that cetacean morbillivirus played a major role. Our findings increase the knowledge on health and disease aspects of this endangered species.
Cetacean morbillivirus (CeMV; family Paramyxoviridae) is a highly infectious pathogen responsible for numerous cetacean mass die-offs worldwide. Currently, there are 3 well-characterized strains (1), the porpoise morbillivirus, the dolphin morbillivirus, and the pilot whale morbillivirus, and 3 less-known strains, including the novel Guiana dolphin strain (GD)–CeMV, recently identified in a single specimen from Brazil (2). CeMV was detected in Ireland, England, and the Netherlands in 1988–1990 (3,4), when the porpoise morbillivirus strain was identified in a small number of stranded harbor porpoises (Phocoena phocoena). Since then, CeMV has been implicated as the causal agent of numerous outbreaks and also endemic, sporadic deaths involving multiple cetacean species throughout the North Sea, north Atlantic Ocean, Mediterranean Sea, Black Sea, Indian Ocean (Western Australia), and Pacific Ocean (Hawaii, Japan, and Australia) (1).
To date, no epizootics linked to CeMV causing the death of large numbers of marine mammals has been detected in the South Atlantic. A Guiana dolphin (Sotalia guianensis) stranded in Espírito Santo, Brazil, which tested positive for CeMV by reverse transcription PCR (RT-PCR) and immunohistochemistry, has been the only confirmed fatal case in South Atlantic cetaceans (2). We describe the results of pathologic and molecular investigations on 20 deceased Guiana dolphins in the onset of the ongoing unusual mortality event in Rio de Janeiro, Brazil.
During November–December 2017, a unusual mortality event involving 56 Guiana dolphins began in Ilha Grande Bay, Rio de Janeiro (Brazil; 23°4′45′′–23°13′38′′S, 44°5′30′′–44°22′28′′W). This area is a relatively well-preserved ecosystem, and Guiana dolphin population census size in this area was estimated at ≈900 animals (5). Stranding occurrence for the same period in previous years ranged from 0 to 3 specimens. During this event, carcasses were recovered adrift or washed ashore. We performed necropsies on 20/56 (37.7%) Guiana dolphins and recorded epidemiologic and biologic data (Table 1).
We collected representative tissue samples of major organs and fixed them in 10% neutral buffered formalin or froze them at −80°C. For PCR analysis, we extracted viral RNA from frozen lung, brain, spleen, liver, and kidney (Table 1) using Brazol Reagent (LGC Biotecnologia Ltda, São Paulo, Brazil), according to the manufacturer’s instructions. We used random primers and M-MLV Reverse Transcription Kit (Invitrogen, Life Technologies, Carlsbad, CA, USA) to synthesize cDNA. We performed amplification using primers targeting highly conserved fragments of the phosphoprotein (P) gene (6) and RNA-dependent RNA polymerase protein coded by the L gene (primers RES-MOR-HEN) as previously described (2,7).

Figure 1. Phylogenetic tree showing partial sequence of A) morbillivirus phosphoprotein and B) large protein genes of cetacean morbillivirus (CeMV) isolates found in stranded Guiana dolphins (Sotalia guianensis) from Rio de Janeiro, Brazil...
We detected CeMV genome in 15/20 (75%) animals for the P gene and 6/6 (100%) animals for the L gene. We sequenced amplified products and compared them with sequences of CeMV using blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi). We obtained identical sequences for the L gene, and 2 sequences with variation in 1 nucleotide position for the P gene. Sequencing of 405-bp amplified fragments of the CeMV P gene revealed 99%–100% identity to GD-CeMV (2) and 78%–82% identity with other CeMV strains. A 443-bp amplified fragment of the CeMV L gene revealed 74%–75% identity to CeMV and other morbillivirus species. Partial P and L gene sequencing and analysis using MEGA7 (http://megasoftware.net/) corroborate that the GD-CeMV strain differed from other morbilliviruses and represented a distinct lineage (Figure 1).
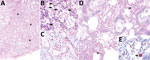
Figure 2. Cetacean morbillivirus–associated histopathologic findings in 2 Guiana dolphins (Sotalia guianensis), a female adult (case 1, panels A–C) and a male calf (case 2, panels D–E). A) The mammary gland parenchyma is...
For histologic examination, we embedded formalin-fixed tissues in paraffin wax, processed them as routine, and stained them with hematoxylin and eosin. We recorded detailed histopathologic findings of 6 animals positive for CeMV by RT-PCR (Table 2). One specimen had lesions consistent with CeMV infection, including marked multifocal, subacute bronchointerstitial pneumonia with type II pneumocyte hyperplasia, syncytia, and scattered intraepithelial, intranuclear, and intracytoplasmic inclusion bodies (INCIBs); mild to moderate multifocal histiocytic and lymphoplasmacytic mastitis with necrosis and epithelial INCIBs (Figure 2, panels A–C); and multicentric lymphoid depletion. In addition, most animals had moderate to severe verminous bronchopneumonia and pleuritis with morphologic evidence of pulmonary arterial hypertension, multicentric eosinophilic and necrotizing lymphadenitis, and chronic aortic endarteritis by adult nematodes and pulmonary endarteritis by migrating larval nematodes histomorphologically compatible with Halocercus brasiliensis (8). Other common findings included moderate to poor body condition and lack of ingesta with small amounts of feces. Two (10%) of the 20 animals (which were negative for CeMV by RT-PCR) showed typical external net markings and multiorgan acute hemodynamic alterations (congestion, edema, and hemorrhage) supporting asphyxia due to bycatch as the cause of death.
We performed immunohistochemistry studies using a monoclonal antibody against the nucleoprotein antigen of canine distemper virus (CDV-NP mAb; VMRD Inc. Pullman, WA, USA), as described (2). In lung tissue sections (cases 1, 2, 11, and 13), we evaluated number and distribution of immunopositive cells and immunolabeling intensity. Lung samples from all animals tested showed widespread and intense immunolabeling in bronchial, bronchiolar, and alveolar epithelium, alveolar macrophages, and syncytia (Figure 2, panels D,E).
In this investigation, typical histopathologic findings consistent with CeMV were evident in 1 animal, indicating a systemic infection. Although chronic bronchointerstitial pneumonia and multicentric lymphoid depletion observed in most animals are common findings in CeMV-infected cetaceans, these lesions were considerably overlapped by H. brasiliensis endoparasitosis. The pathologic signatures of GD-CeMV remain unknown. No other CeMV strain has been described in the South Atlantic Ocean. In subacute and chronic CeMV presentations, fatalities are often ascribed to secondary infections (e.g., toxoplasmosis, aspergillosis) (9,10). In our cohort, autolysis precluded microscopic examinations in some animals, so we could not draw further pathologic conclusions. Nonetheless, moderate to severe parasitosis by H. brasiliensis likely accounted for severe illness in most cases. Intense viral replication in the mammary acinar epithelium in a lactating female may imply a vertical transmission route, in addition to the horizontal aerogenous and direct contact routes (10). Therefore, future pathologic and epidemiologic studies in the South Atlantic should consider vertical transmission. Two cases from this cohort were bycaught, further supporting the multifactorial nature of the ongoing unusual mortality event.
The Guiana dolphin is a coastal and estuarine delphinid endemic from southern Brazil to Central America and one of the most threatened South Atlantic cetaceans, for which recent studies demonstrate severe population decline (11). Because of its near-shore distribution and site fidelity (12), the Guiana dolphin is susceptible to the effects of human activities (e.g., habitat degradation, chemical pollution, noise, and bycatch) (13). Many intricate and complex anthropic and natural factors interplay and modulate the decline of species. Human activities are by far the major threat and cause for decimation of cetacean populations (14); however, natural factors such as highly infectious pathogens, e.g., CeMV, may drive decimating events in susceptible hosts (15).
We provide compelling molecular and pathologic evidence associating GD-CeMV infection with the ongoing Guiana dolphin mass die-off near Rio de Janeiro, Brazil. As of January 2018, this event had resulted in the deaths of >200 Guiana dolphins in southern Rio de Janeiro state, and the deaths appeared to be extending southward. The environmental consequences and conservation effects, coupled with the anthropogenic threats, are expected to be dramatic. The factors underlying the die-off are being investigated, but our results indicate that GD-CeMV plays a major contributory role. Our findings increase the body of knowledge on health and disease aspects of this endangered species.
Dr. Groch is a postdoctoral fellow studying the advancement of pathology of cetaceans in Brazil, particularly of infectious diseases. Her current research focuses on determining geographic and host ranges for CeMV, as well as delineating the pathologic signature and CeMV strains present in cetaceans of Brazil.
Acknowledgments
We thank the Laboratório de Mamíferos Aquáticos e Bioindicadores (MAQUA/UERJ) team for their assistance in stranding monitoring and necropsy procedures and Haydée A. Cunha for helpful comments on the draft of this manuscript.
Programa de Conservação dos botos-cinza (Sotalia guianensis) e outros cetáceos das baías da Ilha Grande e de Sepetiba (MAQUA/UERJ, Associação Cultural e de Pesquisa Noel Rosa, INEA, Transpetro) and Projeto de Monitoramento de Praias da Bacia de Santos (PMP-BS) (MAQUA/UERJ, CTA Meio Ambiente, Instituto Boto Cinza, Petrobras) support cetacean research in this region. PMP-BS is a monitoring program demanded by the federal environmental licensing conducted by IBAMA. This research was also supported by Coordination for the Improvement of Higher Education Personnel (CAPES) and São Paulo Research Foundation (FAPESP), grants #2014/24932-2, #2015/00735-6, and #2017/02223-8. J.L.C. D. is the recipient of a fellowship from the National Research Council (CNPq; grant # 305349/2015-5); A.F.A., J.L-B., and T.L.B. are funded by research grants from CNPq (PQ-1D, PQ-1C, and PQ-2, respectively), FAPERJ (CNE and JCNE, respectively), and UERJ (Prociência).
References
- Van Bressem M-F, Duignan PJ, Banyard A, Barbieri M, Colegrove KM, De Guise S, et al. Cetacean morbillivirus: current knowledge and future directions. Viruses. 2014;6:5145–81. DOIPubMed
- Groch KR, Colosio AC, Marcondes MC, Zucca D, Díaz-Delgado J, Niemeyer C, et al. Novel cetacean morbillivirus in Guiana dolphin, Brazil. Emerg Infect Dis. 2014;20:511–3. DOIPubMed
- Kennedy S, Smyth JA, Cush PF, McCullough SJ, Allan GM, McQuaid S. Viral distemper now found in porpoises. Nature. 1988;336:21. DOIPubMed
- Visser IK, Van Bressem MF, de Swart RL, van de Bildt MW, Vos HW, van der Heijden RW, et al. Characterization of morbilliviruses isolated from dolphins and porpoises in Europe. J Gen Virol. 1993;74:631–41. DOIPubMed
- Souza SCP. Estimation of population parameters of the Guiana dolphin, Sotalia guianensis (Van Bénéden, 1864) (Cetacea, Delphinidae) in Paraty Bay (RJ). Rio de Janeiro (Brazil): Universidade do Estado do Rio de Janeiro; 2013.
- Barrett T, Visser IKG, Mamaev L, Goatley L, van Bressem M-F, Osterhaust AD. Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology. 1993;193:1010–2. DOIPubMed
- Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8. DOIPubMed
- Delyamure SL. Helminthofauna of marine mammals (ecology and phylogeny) [translated from Russian]. Jerusalem: Israel Program for Scientific Translation Ltd.; 1968.
- Stephens N, Duignan PJ, Wang J, Bingham J, Finn H, Bejder L, et al. Cetacean morbillivirus in coastal Indo-Pacific bottlenose dolphins, Western Australia. Emerg Infect Dis. 2014;20:666–70. DOIPubMed
- Domingo M, Visa J, Pumarola M, Marco AJ, Ferrer L, Rabanal R, et al. Pathologic and immunocytochemical studies of morbillivirus infection in striped dolphins (Stenella coeruleoalba). Vet Pathol. 1992;29:1–10. DOIPubMed
- Azevedo AF, Carvalho RR, Kajin M, Van Sluys M, Bisi TL, Cunha HA, et al. The first confirmed decline of a delphinid population from Brazilian waters: 2000–2015 abundance of Sotalia guianensis in Guanabara Bay, South-eastern Brazil. Ecol Indic. 2017;79:1–10. DOI
- Flores PAC, Silva VMFD. Tucuxi and Guiana dolphin: Sotalia fluviatilis and S. guianensis. In: Perrin WF, Wursig B, Thewissen JGM, editors. Encyclopedia of marine mammals, 2nd ed. San Diego (CA): Academic Press; 2009. p. 1188–92.
- Crespo EA, Notarbartolo di Sciara G, Reeves RR, Smith BD. Dolphins, whales, and porpoises: 2002–2010 conservation action plan for the world’s cetaceans. Gland (Switzerland) and Cambridge: International Union for Conservation of Nature; 2003.
- Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. Accelerated modern human-induced species losses: Entering the sixth mass extinction. Sci Adv. 2015;1:e1400253. DOIPubMed
- Forcada J, Aguilar A, Hammond PS, Pastor X, Aguilar R. Distribution and numbers of striped dolphins in the western Mediterranean Sea after the 1990 epizootic outbreak. Mar Mamm Sci. 1994;10:137–50. DOI






















.png)
.png)











No hay comentarios:
Publicar un comentario