
Volume 23, Number 10—October 2017
CME ACTIVITY - Research
Enteric Infections Circulating during Hajj Seasons, 2011–2013
Moataz Abd El Ghany123 , Mona Alsomali1, Malak Almasri1, Eriko Padron Regalado, Raeece Naeem, AbdulHafeez Tukestani, Abdullah Asiri, Grant A. Hill-Cawthorne, Arnab Pain23
, Mona Alsomali1, Malak Almasri1, Eriko Padron Regalado, Raeece Naeem, AbdulHafeez Tukestani, Abdullah Asiri, Grant A. Hill-Cawthorne, Arnab Pain23 , and Ziad A. Memish23
, and Ziad A. Memish23
Abstract
Hajj, the annual Muslim pilgrimage to Mecca, Saudi Arabia, is a unique mass gathering event that raises public health concerns in the host country and globally. Although gastroenteritis and diarrhea are common among Hajj pilgrims, the microbial etiologies of these infections are unknown. We collected 544 fecal samples from pilgrims with medically attended diarrheal illness from 40 countries during the 2011–2013 Hajj seasons and screened the samples for 16 pathogens commonly associated with diarrheal infections. Bacteria were the main agents detected, in 82.9% of the 228 positive samples, followed by viral (6.1%) and parasitic (5.3%) agents. Salmonella spp., Shigella/enteroinvasive Escherichia coli, and enterotoxigenic E. coli were the main pathogens associated with severe symptoms. We identified genes associated with resistance to third-generation cephalosporins ≈40% of Salmonella- and E. coli–positive samples. Hajj-associated foodborne infections pose a major public health risk through the emergence and transmission of antimicrobial drug–resistant bacteria.
Hajj, the annual pilgrimage by Muslims to Mecca, Saudi Arabia, is a unique mass gathering event in terms of scale (i.e., the number of pilgrims), diversity of the pilgrims, nature of the activities performed, and regularity. Approximately 2 million pilgrims from 185 countries, in addition to hundreds of thousands of residents of Saudi Arabia, travel to holy sites in Mecca each year (1). This enormously diverse population (in terms of ethnic origin, socioeconomic status, sex, age, and health status) comes together to perform the same activities within a relatively short period over a limited area of land (2), which allows for the mixing of infectious agents and susceptible populations (3). Mass gatherings such as Hajj therefore increase the potential for the emergence and dissemination of infections and raises public health concerns in Saudi Arabia and globally (4). Hajj-associated communicable public health hazards mainly involve the transmission of respiratory infections, foodborne diseases, bloodborne diseases, and zoonotic infections (4).
Globally, diarrheal infections remain the leading cause of mortality in children <5 years of age and contribute to ≈10% of child deaths each year (5–7). In addition, traveler’s diarrhea is still the most common illness observed in travelers returning from regions where diarrheal diseases are endemic (8,9). The main etiologic agents detected are consistently bacteria (Escherichia coli, Salmonella spp., Shigella spp., and Campylobacter spp.); viruses (rotavirus, norovirus, and adenovirus); and parasites (Cryptosporidium spp., Giardia lamblia, and Entamoeba histolytica) (10,11).
Despite substantial advances in food and water hygiene in many countries, mass gathering events still represent the perfect environments for the transmission of enteric infections (12,13). Diarrheal infections and foodborne diseases are commonly associated with the Hajj pilgrimage (14). Although diarrheal infections and other enteric infections are one of the most common complaints among pilgrims, little information is available regarding incidence, etiologic agents, and the abundance of antimicrobial drug–resistant strains. Published reports have mainly been based on analyses of hospital admission data that lack full characterization of the nature of the infections (15–17). Moreover, estimates of the incidence of Hajj-associated gastrointestinal disease based on hospital admission data can vary considerably (14). Recently, a few studies have shown an increase in the carriage rates of enteric pathogens that include Tropheryma whipplei (18), multidrug-resistant nontyphoidal Salmonella (19), and carbapenemase-producing E. coli (20) among pilgrims from France returning from Hajj. These findings, coupled with the growing threat of drug-resistant microorganisms (21), increase the risks associated with the Hajj pilgrimage and fuels the emergence and dissemination of drug-resistant enteric pathogens.
We conducted a large-scale study to catalog the circulating enteric pathogen population in Hajj pilgrims with diarrheal symptoms. We report on the use of molecular and antigenic approaches to characterize the etiologic agents associated with enteric infections in pilgrims who sought medical treatment while performing Hajj during the 2011–2013 seasons.
Ethics Statement
The samples were originally collected for diagnostic purposes; therefore, collection was not experimental in nature. The Ministry of Health of Saudi Arabia anonymized all identifiable information, and only deidentified records and samples were available to the researchers. The King Fahad Medical City Institutional review board approved the study protocol (approval no. 11–157, dated October 4, 2011). The Institutional Biosafety and Ethics Committee of King Abdullah University of Science and Technology also approved the study in 2013.
Study Design
We conducted the study for 3 successive Hajj seasons, starting in 2011. Fecal samples from pilgrims having medically attended diarrhea while performing Hajj were collected. Healthcare facilities distributed along the Hajj sites were enrolled in the study.
We included patients with symptoms who were seeking medical care for diarrhea or who were admitted to hospitals or primary care centers established in the holy sites during the 7–10 day Hajj period. We defined diarrhea as the occurrence of >3 unformed stools in a 24-hour period or passing stool more frequently than normal for the patient, accompanied with >1 other gastrointestinal symptom (abdominal pain/cramps, vomiting, or bloody or mucoid stools). Patients who had unformed stool with visible blood were defined as having cases of dysentery. Patients with increased body temperature were categorized as having either mild (>37.5°C and <39°C) or severe (>39°C) fever.
We categorized the patients into 2 groups according to degree of symptom severity. We defined severe diarrhea as >6 unformed stools per day; diarrhea requiring hospitalization; or diarrhea accompanied by fever, dehydration, or bloody or mucoid stools. We classified patients with diarrhea not fulfilling the criteria for severe symptoms as having mild cases. We screened all the samples molecularly, antigenically, or both for a panel of 16 infectious agents commonly associated with diarrheal infection.
Antigenic Detection of Viral and Parasitic Pathogens
We used qualitative enzyme immunoassays for the initial detection of viral agents in the fecal samples according to manufacturers’ instructions. We used the IDEIA Norovirus test (Oxoid, Basingstoke, UK) to detect norovirus genogroups 1 and 2 and ProSpecT tests (Oxoid) to detect of group A rotaviruses, adenoviruses, and astroviruses. For parasitic agents, we used the Giardia/Cryptosporidium Quik Chek test (TechLab, Blacksburg VA, USA) for the detection and differentiation of Cryptosporidium oocyst antigen and Giardia cyst antigen.
Isolation of DNA Using QIAsymphony Platform
We used the QIAsymphony SP (QIAGEN, Hilden, Germany), an automated high-throughput platform, for the isolation and purification of total DNA from the collected fecal samples. We used the QIAsymphony DNA 800 complex kit (QIAGEN) to extract DNA from 800 μL of pretreated diluted samples according to the manufacturer’s instructions.
Molecular Characterization of Bacterial Species
We used 3 previously established multiplex PCR assays (M1, 2, and 3) in parallel to detect the bacterial pathogens commonly associated with diarrheal infections (22). The M1 multiplex PCR used primers targeting genes eae and bfpA (enteropathogenic E. coli), aggR (enteroaggregative E. coli) and Vero cytotoxin (enterohemorrhagic E. coli). The M2 multiplex PCR used primers targeting the genes elt and st (enterotoxigenic E. coli [ETEC]), daaE (diffusely adherent E. coli), and virF and ipaH (Shigella spp./enteroinvasive E. coli [EIEC]). The M3 multiplex PCR used primers targeting the hipO gene (Campylobacter jejuni), internal transcribed spacer region (Salmonella spp.), Yersinia stable toxin gene (Yersinia enterocolitica) and rtxA gene (Vibrio cholerae). Primer details and the expected PCR fragment sizes are provided (Technical Appendix[PDF - 415 KB - 4 pages] Table 1). In summary, we mixed 200–400 ng of the extracted total DNA, 1–10 μmol/L of each of the primer pairs, and GoTaq Green Master Mix (Promega, Madison, WI, USA) in a PCR total reaction volume of 25 μL to amplify the target genes. We ran PCR products on a 1.5% agarose electrophoresis gel at 120 volts for 2 hours and identified fragment sizes against positive controls by using the GelPilot 1kb Plus ladder (QIAGEN).
Molecular Characterization of Viral Agents
We used the QIAamp Viral RNA Mini Kit (QIAGEN) to extract viral RNA from antigenically positive samples for rotavirus, norovirus, and astrovirus according to the manufacturer’s instruction. We performed reverse transcription by using the SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA) and PCR amplification by using Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific, Waltham, MA, USA) and previously described primers for the detection of rotavirus (23), norovirus (24), and astrovirus (25). Primer details and expected PCR fragment size are provided (Technical Appendix[PDF - 415 KB - 4 pages] Table 2). We purified PCR products by using the MinElute Gel Extraction kit (QIAGEN); sequencing was performed on an ABI 3730xl (Thermo Fisher Scientific) at the Bioscience Core Laboratory at King Abdullah University of Science and Technology. We used BioEdit Sequence Alignment Editor 7.2.6.1 (http://www.mbio.ncsu.edu/bioedit/page2.html) to trim and align bidirectional sequence reads and used the consensus sequences to identify the viral genotype. We identified rotavirus genotypes by using RotaC version 2.0 software (26) and noroviruses by using genotyping tool version 1.0 (27). We used previously described phylogenetic analyses to identify astrovirus genotypes (28).
Molecular Characterization of β-Lactamase Genes
We further screened the samples positive for 1 of the Enterobacteriaceae species for the detection of β-lactamase genes (blaCTX-M-15, blaIMP, blaKPC, blaNDM, blaOXA-48, and blaVIM) as previously described (29). The list of primers used in the detection of β-lactamase genes and the expected PCR fragment sizes are provided (Technical Appendix[PDF - 415 KB - 4 pages] Table 3).
Statistical Analysis
We evaluated the differences between the sets of the categorical data by using the Pearson χ2 test. We defined statistical significance as p<0.05.
Demographic and Clinical Features of the Patients
During 3 consecutive Hajj seasons (2011–2013), we collected 544 fecal samples from pilgrims who had diarrhea while performing Hajj and who sought treatment at healthcare facilities (Tables 1, 2). These patients originated from 40 countries on 5 continents (Technical Appendix[PDF - 415 KB - 4 pages]Table 4). Most patients (434, 79.8%) originated from 7 countries: Saudi Arabia (24.82%, n = 135), Nigeria (15.07%, n = 82), Egypt (12.87%, n = 70), Bangladesh (8.09%, n = 44), Pakistan (6.43%, n = 35), Yemen (6.25%, n = 34), and India (6.25%, n = 34). Median (+quartile deviation) patient age was 40.17 (+13.17) years. By Hajj season, median age was 40 (+12.25) years in 2011, 40 (+13.25) in 2012, and 40.5 (+14) years in 2013 (Table 1). Most patients were men (72.98%, n = 397); women represented 27.12% of patients in 2011, 28.28% in 2012, and 23.26% in 2013 (Table 1).
We summarized the distribution of the clinical features among the patients during the 3 Hajj seasons (Table 2). Most patients were seen as outpatients (86.95%, n = 473), and the most frequently reported symptoms were abdominal pain/cramp (90.26%, n = 491), presence of mucus in the stool (44.85%, n = 244), watery diarrhea (38.42%, n = 209), dehydration (24.63%, n = 134), vomiting (22.98%, n = 125), and moderate fever (22.06%, n = 120). Less common symptoms were bloody stool (9.38%, n = 51) and severe fever (6.62%, n = 36). We observed significant differences in the frequencies of these symptoms across the 3 Hajj seasons (Table 2).
Characterization of Bacterial Pathogens
We screened the 544 fecal samples collected from the patients during the 2011–2013 Hajj seasons for 16 infectious agents, including bacteria, viruses, and parasites commonly associated with diarrheal infections. We calculated the number of the samples tested and the number and percentage of the positive samples from each season (Table 3). We detected >1 of the pathogens screened for in 41.91% (n = 228) of the samples. We observed no significant difference between the numbers of positive samples during the 3 seasons (χ2 = 0.63; p = 0.73). The percentages of positive samples detected were 43.22% (n = 51) for 2011, 40.40% (n = 120) for 2012, and 44.19% (n = 57) for 2013. Bacterial pathogens were the predominant infectious agents detected for the 3 Hajj seasons and the agents identified in 34.74% (n = 189) of the total samples, followed by viral (2.57%, n = 14) and parasitic (2.21%, n = 12) agents. Thirteen patients (representing 2.39% of the total samples) had samples testing positive for >1 pathogen. We observed no significant difference in the distribution of infectious agents across the 3 seasons (χ2 = 8.84; p = 0.18).
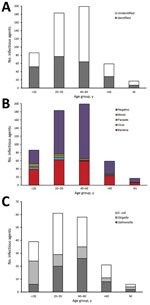
Figure. Distribution of infectious agents among persons who acquired enteric infections during their travel for Hajj, 2011–2013, by age group. A) Identified versus unidentified samples; B) type of pathogen; C) bacterial agent....
We calculated the distribution of patients by age group and the enteric pathogens identified (Figure, panels A, B). The highest proportion of patients having diarrhea of known etiology, compared with unknown, was the <20-year-old age group (odds ratio [OR] 2.46; p = 0.0002). Conversely, the highest proportion of patients having diarrhea of unknown etiology compared with known was the 40–60 years age group (OR 0.52; p = 0.0004). For most of the age groups, bacteria were the main cause of diarrhea in patients, with no significant difference detected across the 3 Hajj seasons (χ2 = 8.59; p = 0.2).
We also calculated the distribution of the bacterial agents associated with the diarrheal patients during 2011–2013 Hajj seasons by age group (Figure, panel C). E. coli was the most frequent species present, detected in 43.39% (n = 82) of the bacteria-positive samples. Of the serovars tested, ETEC was the most common, detected in 25.4% (n = 48) of the positive samples, followed by enteropathogenic E. coli (8.47%, n = 16), enteroaggregative E. coli (3.7%, n = 7), diffusely adherent E. coli (3.7%, n = 7), and enterohemorrhagic E. coli (2.12%, n = 4). We detected Salmonella spp. in 32.80% (n = 62) and Shigella/EIEC in 21.69% (n = 41) of the bacteria-positive samples. We observed significant differences in the distribution of bacterial pathogens across the 3 Hajj seasons (χ2 = 12.89; p = 0.01) and among the different age groups (χ2 = 21.62; p = 0.01).
Characterization of Viral and Parasitic Pathogens
We calculated the distribution of the viral and parasitic agents associated with diarrheal infections of pilgrims during the 2011–2013 Hajj seasons (Table 3). Screening for adenoviruses, astroviruses, noroviruses, and rotaviruses showed rotaviruses were most common, detected in 42.86% (n = 6) of the samples positive for the screened viruses. Astroviruses were detected in 21.43% (n = 3), noroviruses in 28.57% (n = 4), and adenoviruses in 7.14% (n = 1) of the virus-positive samples. We used reverse transcription PCR and Sanger sequencing to determine the genotypes of the astroviruses, noroviruses, and rotaviruses detected (Technical Appendix[PDF - 415 KB - 4 pages] Table 5). All norovirus genotypes identified were recovered from pilgrims from inside Saudi Arabia. Also, 80% of the identified astrovirus genotypes were recovered only from pilgrims from inside Saudi Arabia (astrovirus 2 or 5), whereas the single astrovirus 1 genotype was recovered from a pilgrim from Morocco (Technical Appendix[PDF - 415 KB - 4 pages] Table 5).
Giardia spp. was the most common parasitic agent, identified in 83.33% (n = 10) of the parasite-positive samples, followed by Cryptosporidium spp. in 16.66% (n = 2) of the samples. We isolated Giardia spp. from patients originating from 10 countries: 4 from Pakistan, 3 from Nigeria, 2 from Bangladesh, and 1 each from Ethiopia, Somalia, Egypt, Jordan, Niger, India, and Afghanistan. We identified Cryptosporidium spp. in 2 children (<5 years of age) from Saudi Arabia and 1 older pilgrim (65 years of age) from Chad (Technical Appendix[PDF - 415 KB - 4 pages] Table 6).
Relationship between Severity of Diarrheal Disease and Etiologic Agent
We calculated the distribution of the etiologic agents by severity of disease (Table 4). The percentage of samples with identified etiologic agents was significantly higher in patients with severe cases compared with those with mild cases (OR 1.69; p = 0.01). Similarly, the percentage of bacterial agents was significantly higher in patients with severe cases compared with those with mild cases (OR 1.58; p = 0.04). The main bacterial contributors to the severe disease of Hajj-associated diarrheal illness were Salmonella, Shigella/EIEC, and ETEC.
Antimicrobial Drug Resistance
We calculated the distribution of β-lactamase genes among the identified bacterial samples (Table 5). blaCTX-M-15 and blaNDM were the most common antimicrobial resistance genes, associated predominantly with Salmonella (n = 25/62) and ETEC (n = 16/48). This finding suggests that 40.32% of Salmonella infections and 33.33% ETEC infections associated with the Hajj might be resistant to at least some third-generation cephalosporins, and this number might be growing with successive seasons.
Enteric infections are commonly associated with mass gathering events, including the annual Hajj pilgrimage to Mecca, Saudi Arabia. The host country and the country of origin of many of the pilgrims are endemic for enteric pathogens and increasingly high levels of antimicrobial resistance. In addition, the lack of effective vaccines against major bacterial infections is challenging (30). These circumstances raise serious public health challenges for Saudi Arabia, with potential intercontinental and global implications. A key challenge is the paucity of information available on the structure of the circulating enteric pathogens during Hajj. Comprehensive information on the etiologic agents associated with Hajj-associated diarrheal disease is lacking (14). Recent studies have found increased rates of carriage of multidrug-resistant bacteria, including Salmonella spp., (19) E. coli (20), and Acinetobacter baumannii (20) in pilgrims returning home to France after performing Hajj. However, these studies have only focused on colonization by antimicrobial-resistant bacteria in a particular host population.
In this study, we used integrated antigenic and molecular approaches to screen 544 fecal samples from pilgrims with medically attended diarrheal illness for 16 pathogens to identify the etiologic agents responsible for patients seeking care at healthcare facilities during 3 consecutive Hajj seasons. Bacterial pathogens were the most common causes of Hajj-associated diarrheal disease, followed by viruses and parasites, and this pattern was maintained during all 3 seasons.
Our data demonstrate that Hajj-associated diarrheal disease is usually caused by 1 bacterial agent, with ETEC, Salmonella spp., and Shigella/EIEC being the most common. This association is distinct to the pattern of travelers’ diarrhea observed in travelers from Finland, where multiple bacterial pathogens have been identified in 53% of patients with ongoing diarrhea and 25% of those without symptoms (31). However, this observation is not surprising; Hajj-associated diarrheal disease is likely to be different from travelers’ diarrhea because of the different populations involved. Most of Hajj pilgrims originate from intermediate- and high-risk regions for enteric pathogens. In contrast, many travelers’ diarrhea patients are nonimmune persons from developed countries who are naive to many of the enteric pathogens encountered and thus are more highly susceptible to infection when traveling overseas (32).
Viruses ranked second and parasites third as the most commonly detected pathogens in patients with Hajj-associated diarrhea. Of note, all of the identified noroviruses and most astroviruses and rotaviruses were recovered from pilgrims from inside Saudi Arabia. The emergent norovirus genotype GII.4 that was first identified in Sydney, Australia, in 2012 and subsequently resulted in global outbreaks had already begun circulating among pilgrims from Saudi Arabia in late October and early November of the 2012 Hajj season. Major causes of diarrhea among children living in Saudi Arabia include rotaviruses (accounting for 6.0% incidence), noroviruses (3.5%), astroviruses (1.9%), and adenoviruses (1.4%) (33).
The 3 most commonly identified bacteria in our study (Salmonella spp., Shigella spp., and E. coli) have all been identified by the World Health Organization as being among the top 9 bacteria likely to have a serious impact on global public health (21). Of particular concern were the presence of extended-spectrum β-lactamase (ESBL) (primarily blaCTX-M-15) and carbapenemase (e.g., blaNDM) genes in ≈40% of Salmonella spp. and E. coli–positive samples collected.
Recently, travelers’ diarrhea has been shown to be an independent risk factor for contracting ESBL-producing Enterobacteriaceae (ESBL-PE) but not carbapenemase-producing Enterobacteriaceae (CPE), with the rate of acquisition varying by destination (34). Saudi Arabia and the countries of origin for many of the pilgrims are countries at high risk for the acquisition of diarrheal (9,32) and ESBL-PE infections (34). Recent surveillance studies have also reported increasing prevalence of CPE and ESBL-PE isolates in the Gulf Cooperation Council countries (35), with some research institutes in Saudi Arabia finding that up to 65% of E. coli isolates are ESBL producers (36). Recently, the rates of blaCTX-M-15 infection in Hajj pilgrims have been found to be 31% in 2013 and 34.83% in 2014 (37).
Collectively, these results suggest that further epidemiologic investigations need to be carried out during pilgrimages to identify potential food sources of pilgrim infections. In addition, antimicrobial drug susceptibility testing is needed to inform treatment.
This study used a retrospective approach and 1 anonymized specimen from each patient enrolled in the study. One advantage of this approach is that the study population is more representative of the highly diverse Hajj population, with samples collected from patients originating from 40 different countries. However, a prospective approach with pre- and post-Hajj samples collected from each patient would have provided information on the role of the pilgrimage in contracting the pathogens identified.
In addition, even though integrated molecular and antigenic approaches were used, >50% of the tested samples had no identifiable etiologic agent. These samples require further examination using more comprehensive high-throughput sequencing and metagenomic approaches. High-throughput shotgun sequencing has been used successfully to study population structures and define the epidemiologic links of many enteric pathogens (38–42). Moreover, metagenomic approaches have been used successfully to identify viral (43,44) and bacterial (45) agents associated with enteric infections. This approach could enable estimation of the ratio of pathogenic to commensal bacteria in pilgrims’ guts, thereby characterizing the acquisition of potential pathogens and their dynamics before and during infections.
Finally, in this study, the assessment of antimicrobial drug susceptibility was only performed by detecting resistance-related genes. The presence of such genes does not necessarily mean the pathogen identified is carrying them, and these genes might be associated with other commensal carriage. We focused on those resistance genes that are posing the most risk to global health and can be easily shared among the Enterobacteriaceae, rather than the genes that can confer resistance to the antibiotics widely used for treating enteric infections.
The data we have collected are alarming and highlight the need for further studies to explore the impact of Hajj on public health in Saudi Arabia and globally. Longitudinal studies are required to monitor the changes in colonization patterns of pilgrims during the Hajj, identify the key factors that control these changes, detect the emergence of novel variants (particularly those associated with drug resistance), and understand the dynamics of disease transmission. In addition, active surveillance for enteric diseases is needed to define the potential impact of Hajj on the baseline status of enteric infections in residents of Saudi Arabia and to investigate foodborne outbreaks of disease in a timely manner.
Dr. Abd El Ghany is a senior researcher at the Westmead Institute for Medical Research and the Marie Bashir Institute for Infectious Diseases and Biosecurity, University of Sydney, Sydney, Australia. His primary research interests are the use of molecular and different Omics approaches (i.e., whole genome, whole transcriptome, functional genomic, and whole genome sequence–based metagenomic technologies) to address the emergence and transmission of infectious diseases, with particular focus on antimicrobial resistance, food security, and mass gathering medicine.
Acknowledgments
We thank Elizabeth Barnes, Mahmoud Diaf, and Moustafa El Badry for their advice regarding the statistical analysis.
This work was supported by the Saudi Ministry of Health (Z.M.) and King Abdullah University of Science and Technology (faculty baseline funding [BAS/1/1020-01-01] to A.P.), and Marie Bashir Institute and Sydney Medical School Foundation (M.A. and G.H.).
References
- Memish ZA, Venkatesh S, Ahmed QA. Travel epidemiology: the Saudi perspective. Int J Antimicrob Agents. 2003;21:96–101. DOIPubMed
- Memish ZA, Stephens GM, Steffen R, Ahmed QA. Emergence of medicine for mass gatherings: lessons from the Hajj. Lancet Infect Dis. 2012;12:56–65. DOIPubMed
- Abubakar I, Gautret P, Brunette GW, Blumberg L, Johnson D, Poumerol G, et al. Global perspectives for prevention of infectious diseases associated with mass gatherings. Lancet Infect Dis. 2012;12:66–74. DOIPubMed
- Ahmed QA, Arabi YM, Memish ZA. Health risks at the Hajj. Lancet. 2006;367:1008–15. DOIPubMed
- GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–71. DOIPubMed
- Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al.; Child Health Epidemiology Reference Group of WHO and UNICEF. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379:2151–61. DOIPubMed
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22. DOIPubMed
- Steffen R. Epidemiology of travellers’ diarrhea. J Travel Med. 2017;24(suppl_1):S2–5. DOIPubMed
- Steffen R, Hill DR, DuPont HL. Traveler’s diarrhea: a clinical review. JAMA. 2015;313:71–80. DOIPubMed
- Diemert DJ. Prevention and self-treatment of traveler’s diarrhea. Clin Microbiol Rev. 2006;19:583–94. DOIPubMed
- Barrett J, Brown M. Travellers’ diarrhoea. BMJ. 2016;353:i1937. DOIPubMed
- Botelho-Nevers E, Gautret P. Outbreaks associated to large open air festivals, including music festivals, 1980 to 2012. Euro Surveill. 2013;18:20426.PubMed
- Gautret P, Steffen R. Communicable diseases as health risks at mass gatherings other than Hajj: what is the evidence? Int J Infect Dis. 2016;47:46–52. DOIPubMed
- Gautret P, Benkouiten S, Sridhar S, Al-Tawfiq JA, Memish ZA. Diarrhea at the Hajj and Umrah. Travel Med Infect Dis. 2015;13:159–66. DOIPubMed
- Al-Ghamdi SM, Akbar HO, Qari YA, Fathaldin OA, Al-Rashed RS. Pattern of admission to hospitals during muslim pilgrimage (Hajj). Saudi Med J. 2003;24:1073–6.PubMed
- Bakhsh AR, Sindy AI, Baljoon MJ, Dhafar KO, Gazzaz ZJ, Baig M, et al. Diseases pattern among patients attending Holy Mosque (Haram) Medical Centers during Hajj 1434 (2013). Saudi Med J. 2015;36:962–6. DOIPubMed
- Khan NA, Ishag AM, Ahmad MS, El-Sayed FM, Bachal ZA, Abbas TG. Pattern of medical diseases and determinants of prognosis of hospitalization during 2005 Muslim pilgrimage Hajj in a tertiary care hospital. A prospective cohort study. Saudi Med J. 2006;27:1373–80.PubMed
- Gautret P, Benkouiten S, Parola P, Brouqui P, Memish Z, Raoult D. Occurrence of Tropheryma whipplei during diarrhea in Hajj pilgrims: a PCR analysis of paired rectal swabs. Travel Med Infect Dis. 2014;12:481–4. DOIPubMed
- Olaitan AO, Dia NM, Gautret P, Benkouiten S, Belhouchat K, Drali T, et al. Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj. Int J Antimicrob Agents. 2015;45:600–4. DOIPubMed
- Leangapichart T, Gautret P, Griffiths K, Belhouchat K, Memish Z, Raoult D, et al. Acquisition of a high diversity of bacteria during the Hajj pilgrimage, including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 carbapenemase genes. Antimicrob Agents Chemother. 2016;60:5942–8. DOIPubMed
- World Health Organization. Antimicrobial resistance: global report on surveillance 2014 [cited 2016 Oct 1]. http://www.who.int/drugresistance/documents/surveillancereport
- Gómez-Duarte OG, Bai J, Newell E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;63:1–9. DOIPubMed
- Tamura T, Nishikawa M, Anh DD, Suzuki H. Molecular epidemiological study of rotavirus and norovirus infections among children with acute gastroenteritis in Nha Trang, Vietnam, December 2005-June 2006. Jpn J Infect Dis. 2010;63:405–11.PubMed
- Mattison K, Grudeski E, Auk B, Charest H, Drews SJ, Fritzinger A, et al. Multicenter comparison of two norovirus ORF2-based genotyping protocols.J Clin Microbiol. 2009;47:3927–32. DOIPubMed
- Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol. 1995;33:797–801.PubMed
- Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009;9:238. DOIPubMed
- Kroneman A, Vennema H, Deforche K, v d Avoort H, Peñaranda S, Oberste MS, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–5. DOIPubMed
- Guix S, Caballero S, Villena C, Bartolomé R, Latorre C, Rabella N, et al. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J Clin Microbiol. 2002;40:133–9. DOIPubMed
- Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014;58:3085–90. DOIPubMed
- Abd El Ghany M, Sharaf H, Hill-Cawthorne GA. Hajj vaccinations-facts, challenges, and hope. Int J Infect Dis. 2016;47:29–37. DOIPubMed
- Lääveri T, Antikainen J, Pakkanen SH, Kirveskari J, Kantele A. Prospective study of pathogens in asymptomatic travellers and those with diarrhoea: aetiological agents revisited. Clin Microbiol Infect. 2016;22:535–41. DOIPubMed
- Steffen R. Epidemiology of traveler’s diarrhea. Clin Infect Dis. 2005;41(Suppl 8):S536–40. DOIPubMed
- Tayeb HT, Dela Cruz DM, Al-Qahtani A, Al-Ahdal MN, Carter MJ. Enteric viruses in pediatric diarrhea in Saudi Arabia. J Med Virol. 2008;80:1919–29. DOIPubMed
- Kantele A, Lääveri T, Mero S, Vilkman K, Pakkanen SH, Ollgren J, et al. Antimicrobials increase travelers’ risk of colonization by extended-spectrum betalactamase-producing Enterobacteriaceae. Clin Infect Dis. 2015;60:837–46. DOIPubMed
- Balkhy HH, Assiri AM, Mousa HA, Al-Abri SS, Al-Katheeri H, Alansari H, et al.; at the workshop. The strategic plan for combating antimicrobial resistance in Gulf Cooperation Council States. J Infect Public Health. 2016;9:375–85. DOIPubMed
- Zowawi HM. Antimicrobial resistance in Saudi Arabia. An urgent call for an immediate action. Saudi Med J. 2016;37:935–40. DOIPubMed
- Leangapichart T, Tissot-Dupont H, Raoult D, Memish ZA, Rolain JM, Gautret P. Risk factors for acquisition of CTX-M genes in pilgrims during Hajj 2013 and 2014. J Antimicrob Chemother. 2017;72:2627–35; [Epub ahead of print].PubMed
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. DOIPubMed
- Abd El Ghany M, Chander J, Mutreja A, Rashid M, Hill-Cawthorne GA, Ali S, et al. The population structure of Vibrio cholerae from the Chandigarh Region of Northern India. PLoS Negl Trop Dis. 2014;8:e2981. DOIPubMed
- Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–9. DOIPubMed
- Holt KE, Baker S, Weill FX, Holmes EC, Kitchen A, Yu J, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44:1056–9. DOIPubMed
- von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, et al. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet. 2014;46:1321–6. DOIPubMed
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. DOIPubMed
- Nakamura S, Yang CS, Sakon N, Ueda M, Tougan T, Yamashita A, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. DOIPubMed
- Nakamura S, Maeda N, Miron IM, Yoh M, Izutsu K, Kataoka C, et al. Metagenomic diagnosis of bacterial infections. Emerg Infect Dis. 2008;14:1784–6. DOIPubMed






















.png)












No hay comentarios:
Publicar un comentario