
Volume 26, Number 7—July 2020
Dispatch
Shuni Virus in Wildlife and Nonequine Domestic Animals, South Africa
On This Page
Article Metrics
Jumari Steyn, Pebetsi Motlou, Charmaine van Eeden, Marthi Pretorius, Voula I. Stivaktas, June Williams, Louwtjie P. Snyman, Peter E. Buss, Brianna Beechler, Anna Jolles, Eva Perez-Martin, Jan G. Myburgh, Johan Steyl, and Marietjie Venter
Abstract
We screened nonequine animals with unexplained neurologic signs or death in South Africa during 2010–2018 for Shuni virus (SHUV). SHUV was detected in 3.3% of wildlife, 1.1% of domestic, and 2.0% of avian species. Seropositivity was also demonstrated in wildlife. These results suggest a range of possible SHUV hosts in Africa.
Shuni virus (SHUV) (Peribunyaviridae: Orthobunyavirus) was isolated in the 1960s from livestock, Culicoides midges, and a febrile child in Nigeria (1,2). In South Africa, SHUV was identified as the causative agent of neurologic disease in horses (3); seropositivity was also demonstrated in 3.0% of veterinarians, suggesting human exposures (4). SHUV was subsequently identified in aborted livestock and cattle with neurologic disease in Israel, suggesting an extended range beyond Africa (5,6). We investigated other potential susceptible species in South Africa.
During February 2010–September 2018, a total of 101 whole blood, 71 serum, and 476 tissue specimens from 608 nonequine domestic animals, wildlife, and birds (19 fetuses, 118 juvenile and 471 adults) with unexplained neurologic or febrile disease or sudden unexplained death from across South Africa were submitted to the Zoonotic Arbo and Respiratory Virus Program, Centre for Viral Zoonoses, University of Pretoria (Pretoria, South Africa) as part of a passive zoonotic arbovirus surveillance program. We extracted RNA under Biosafety Level (BSL) 3 conditions using the QIAamp viral RNA mini kit (blood) or the RNeasy mini kit (tissue) (QIAGEN, ), according to the manufacturer’s recommendations. We screened all samples by a SHUV real-time reverse transcription PCR (rRT-PCR) (7) and a newly designed rRT-PCR targeting a conserved area of the S segment of the Simbu serogroup (Appendix). We confirmed PCR-positive samples by Sanger sequencing (Inqaba Biotech, ) and phylogenetic analyses (Appendix). We also screened all specimens for West Nile (WNV), Wesselsbron (8), Middelburg (MIDV), Sindbis (9), and equine encephalosis viruses (10).
In addition, serum samples from African buffalo (Syncerus caffer) (n = 45) and white rhinoceros (Ceratotherium simum) (n = 48) from Kruger National Park were collected in March 2014 and June 2016, respectively, by South African National Parks and from wild Nile crocodiles (Crocodylus niloticus) (n = 34) from northern KwaZulu-Natal collected during 2009–2012 by the Faculty of Veterinary Sciences, University of Pretoria, as part of surveillance studies. We examined tissue samples from a SHUV PCR-confirmed positive buffalo (MVA43/10) microscopically under a light microscope using routinely prepared hematoxylin and eosin stained (11) histological sections at the Section of Pathology, Department of Paraclinical Sciences, Faculty of Veterinary Science, University of Pretoria. We subjected serum samples to an epitope-blocking ELISA (eb-ELISA) (12,13) with modifications to detect antibodies to SHUV (Appendix).
We calculated odds ratios (OR) and 95% CI in EpiInfo version 7.2.0.1 (https://www.cdc.gov/epiinfo/index.html). We excluded animals that were found dead, aborted, or stillborn from OR analysis.
We detected SHUV RNA in 15/608 (2.5%) animals tested from 10 different animal species: 12/361 (3.3%) wildlife, 2/196 (1.0%) nonequine domestic animals, and 1/51 avian species (2.0%) (Table 1). We detected SHUV in samples submitted from 2/62 (3.2%) white rhinoceroses, 2/50 (4.0%) sables, 1/15 (6.7%) warthog, 4/54 (7.4%) buffalo, 1/12 (8.3%) crocodiles, 1/5 (20.0%) giraffes, 1/4 (25.0%) springboks, 1/93 (1.1%) domestic bovids, and 1/10 (10.0%) alpacas (Table 1). We also detected SHUV in an exotic monal pheasant (1/13, 7.7%). Differential screening revealed co-infections with MIDV and WNV, suggesting that these arboviruses could co-circulate. A sable was also co-infected with Theileria sp. sable and Theileria separate (Table 1).
In 9/15 (60.0%, 95% CI 35.2%–84.8%) positive infections, we detected SHUV in the central nervous system (CNS) (Table 1), indicating passage across the blood–brain barrier, which suggests SHUV as the likely causal agent of the observed neurologic signs. This finding suggests that SHUV is not just an agent of subclinical infections or reproductive problems, such as abortion, as previously reported (5,14), but is also the likely etiology for neurologic disease in these species, as previously described for horses (3) and cattle (6). We did not detect SHUV RNA in aborted (n = 24) or stillborn (n = 16) animals. Eleven SHUV-positive animals showed neurologic signs (OR 1.8, 95% CI 0.2–14.4), with 2 animals also reported to be pyrexic (OR 2.0, 95% CI 0.4–9.4) or showing respiratory signs (OR 1.0, 95% CI 0.2–4.8) (Table 2). Three SHUV-positive animals were found dead (OR 1.8, 95% CI 0.5–6.4) (Table 2). Specific neurologic signs associated with SHUV infection included hind limb paresis progressing to quadriparesis with normal mentation (OR 6.7, 95% CI 2.0–22.5) (Table 2).
Positivity of infection was highest in the North West (4/47, 8.5% of samples submitted from North West), followed by Limpopo Province (8/132, 6.1%) (Table 1; Appendix Figure 1). SHUV was detected only in 2010 (4/15, 26.7%), 2017 (2/15, 13.3%), and 2018 (9/15, 60.0%) despite continuous surveillance throughout the years, suggesting that outbreaks may be sporadic rather than annual. SHUV PCR positives were detected during April–September in each of the 3 years (Appendix Figure 2).
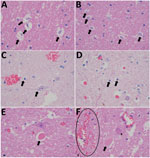
Necropsy examination on the buffalo showed no specific macroscopic lesions on histopathology examination of brain tissue (Figure 1). Pathological changes that could be detected in regions of the brain included mild white matter cerebro–cerebellar gliosis, especially microglial, associated with considerable glial apoptotic activity and occasional perivascular hemorrhage. In the spinal cord, occasional single neuronal necrosis (chromatolysis) and perineuronal hypereosinophilic bodies affecting the dorsal horns of the gray matter were distinctive. This finding seemed to be most severe in the lumbar spinal region. No evidence of demyelination or major immunological reaction was observed, apart from occasional perivascular lymphocytes. Development of appropriate antibodies for immunohistochemistry or probes for in situ hybridization may further describe the pathology of SHUV in animal tissue.
We used phylogenetic analyses on the small segment of the Simbu serogroup to verify the molecular results. All novel sequences from this study, with 1 exception, were closely related to SHUV strains identified in horses in South Africa (3) in clade 1a of lineage I within the Simbu serogroup (bootstopping: posterior probabilities = 89:0.99) (Figure 2). An isolate from a springbok (ZRU261_17_3) clustered with Sango virus (bootstopping: posterior probabilities = 67:0.94). P-distance analysis based on the partial small segment demonstrated few nucleotide differences between novel SHUV strains and reference strains (98.0%–100.0% identity). Wildlife specimens were submitted mostly from dead animals that were already undergoing postmortem cytolysis, inhibiting further genetic analysis and isolation of the virus. The use of a PCR designed to detect Simbu group/orthobunyavirus genus PCR rather than SHUV-specific PCR facilitated detection of these infections.
We detected antibodies to SHUV by an eb-ELISA in 3/44 (6.8%) African buffalo and 2/48 (4.2%) white rhinoceroses but none in crocodiles. SHUV-specific IgG was confirmed, using microtiter virus neutralization assay, in 1 buffalo and 1 rhinoceros. Two of 3 buffalo and 1 rhinoceros positive for SHUV epitope antibodies were negative by microtiter virus neutralization assay, suggesting that these antibodies may have been elicited in response to closely related orthobunyavirus. Confirmation for the third buffalo was not possible because of depleted serum.
Our findings suggest that SHUV may have a wide host range, including several wildlife and domestic species, and should be included in the differential diagnosis of neurologic disease in animals. This study highlights the role of this virus as a potential emerging zoonotic pathogen in Africa that warrants increased surveillance and further investigation. Future epidemiologic studies would benefit from an increased sample size and more extensive serosurveys. Investigation of human infections may define SHUV’s importance as a zoonosis. The causative link between clinical manifestations in the various species and the evidence of SHUV infection must be regarded with caution because other possible infectious and noninfectious etiologies were not excluded by comprehensive investigations in all cases.
Dr. Steyn is a virologist and PhD candidate at the Centre for Viral Zoonoses at the University of Pretoria, South Africa. Her primary research focuses on investigating arboviruses with zoonotic potential at human–animal interface areas.
Acknowledgments
We thank participating veterinarians and veterinary pathologists from South Africa, as well as L.P. Snyman, for the help with the phylogenetic analyses.
This research was funded through the US CDC Global Disease Detection grant for Zoonotic arboviruses under grant no. 1U19GH000571-01-GDD Non-Research CoAg with the NHLS project 23 and UP Zoonotic Arbo and Respiratory virus program, 2012–2015, and by cooperative agreement no. 5 NU2GGH001874-02-00 with the University of Pretoria, 2015–2016. Buffalo samples were financed through BBSRC-USDA funding (grant no. BB/L011085/1) and transported under a Red Cross permit (LDK2016/9/1) to the BSL3 lab at CVZ. J Steyn received doctoral scholarship form the NRF (grant no. 95175), the Meat Industry Trust (grant no. IT8114/98), the Poliomyelitis Research Foundation (grant no. 15/112).
This study was cleared by section 20 (12/11/1/1) approval through the Department of Agriculture Forestry and Fisheries (DAFF) (2014-October 2018) (M.V.) as well as by the animal ethics (H12-16) and section 20 (V057-15)(2017) of the University of Pretoria and the PhD research committee.
References
- Causey OR, Kemp GE, Causey CE, Lee VH. Isolations of Simbu-group viruses in Ibadan, Nigeria 1964-69, including the new types Sango, Shamonda, Sabo and Shuni. Ann Trop Med Parasitol. 1972;66:357–62.
- Lee VH. Isolation of viruses from field populations of culicoides (Diptera: Ceratopogonidae) in Nigeria. J Med Entomol. 1979;16:76–9.
- van Eeden C, Williams JH, Gerdes TGH, van Wilpe E, Viljoen A, Swanepoel R, et al. Shuni virus as cause of neurologic disease in horses. Emerg Infect Dis. 2012;18:318–21.
- van Eeden C, Swanepoel R, Venter M. Antibodies against West Nile and Shuni viruses in veterinarians, South Africa. Emerg Infect Dis. 2014;20:1409–11.
- Golender N, Brenner J, Valdman M, Khinich Y, Bumbarov V, Panshin A, et al. Malformations caused by Shuni virus in ruminants, Israel, 2014–2015. Emerg Infect Dis. 2015;21:2267–8.
- Golender N, Bumbarov V, Assis I, Beer M, Khinich Y, Koren O, Edery N, Eldar A, et al. Shuni virus in Israel: neurological disease and fatalities in cattle. Transbound Emerg Dis. 2019;66:1126–31.
- Van Eeden C, Zaayman D, Venter M. A sensitive nested real-time RT-PCR for the detection of Shuni virus. J Virol Methods. 2014;195:100–5.
- Zaayman D, Human S, Venter M. A highly sensitive method for the detection and genotyping of West Nile virus by real-time PCR. J Virol Methods. 2009;157:155–60.
- van Niekerk S, Human S, Williams J, van Wilpe E, Pretorius M, Swanepoel R, et al. Sindbis and Middelburg old world alphaviruses associated with neurologic disease in horses, South Africa. Emerg Infect Dis. 2015;21:2225–9.
- van Niekerk M, Freeman M, Paweska JT, Howell PG, Guthrie AJ, Potgieter AC, et al. Variation in the NS3 gene and protein in South African isolates of bluetongue and equine encephalosis viruses. J Gen Virol. 2003;84:581–90.
- Bancroft JD, Gamble M, editors. Theory and practice of histological techniques. Edinburgh: Churchill Livingstone; 2002.
- Blitvich BJ, Marlenee NL, Hall RA, Calisher CH, Bowen RA, Roehrig JT, et al. Epitope-blocking enzyme-linked immunosorbent assays for the detection of serum antibodies to west nile virus in multiple avian species. J Clin Microbiol. 2003a;41:1041–7.
- Blitvich BJ, Bowen RA, Marlenee NL, Hall RA, Bunning ML, Beaty BJ. Epitope-blocking enzyme-linked immunosorbent assays for detection of west nile virus antibodies in domestic mammals. J Clin Microbiol. 2003b;41:2676–9.
- Mathew C, Klevar S, Elbers ARW, van der Poel WHM, Kirkland PD, Godfroid J, et al. Detection of serum neutralizing antibodies to Simbu sero-group viruses in cattle in Tanzania. BMC Vet Res. 2015;11:208.
- Saeed MF, Li L, Wang H, Weaver SC, Barrett ADT. Phylogeny of the Simbu serogroup of the genus Bunyavirus. J Gen Virol. 2001;82:2173–81.
Figures
Tables
Cite This ArticleOriginal Publication Date: June 11, 2020






















.png)












No hay comentarios:
Publicar un comentario