
Volume 26, Number 5—May 2020
Research Letter
Serologic Detection of Middle East Respiratory Syndrome Coronavirus Functional Antibodies
On This Page
Figures
Article Metrics
Nisreen M.A. Okba, Ivy Widjaja, Wentao Li, Corine H. GeurtsvanKessel, Elmoubasher A.B.A. Farag, Mohammed Al-Hajri, Wan Beom Park, Myoung-don Oh, Chantal B.E.M. Reusken, Marion P.G. Koopmans, Berend-Jan Bosch , and Bart L. Haagmans
, and Bart L. Haagmans
Abstract
We developed and validated 2 species-independent protein-based assays to detect Middle East respiratory syndrome coronavirus functional antibodies that can block virus receptor-binding or sialic acid-attachment. Antibody levels measured in both assays correlated strongly with virus-neutralizing antibody titers, proving their use for serologic confirmatory diagnosis of Middle East respiratory syndrome.
The zoonotic introductions and ongoing outbreaks of Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) pose a global threat (1,2) necessitating continuous serosurveillance to monitor virus spread alongside the development of vaccine and antibodies as countermeasures. Both approaches require validated assays to evaluate specific antibody responses. Although MERS-CoV serologic assays have been developed (2–6), those detecting functional antibodies cannot be applied in all laboratories and can require Biosafety Level 3 (BSL-3) containment. Recombinant protein-based immunoassays are easier to operate and standardize and do not require BSL-3 containment. However, MERS-CoV protein-based assays developed thus far can only detect antibody binding and give no information on antibody functionality. The MERS-CoV spike protein N terminal subunit (S1) contains 2 functional domains: the N-terminal domain (S1A), which binds sialic acid, the viral attachment factor; and the receptor-binding domain (RBD) (S1B), which binds dipeptidyl peptidase 4, the virus receptor (7,8). Antibodies against those 2 domains can block MERS-CoV infection (9). Based on this fundamental knowledge, we developed 2 recombinant protein-based functional assays.
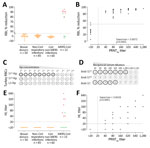
First, we developed an S1-based competitive ELISA, a receptor-binding inhibition assay (RBI), to test for antibodies that block the interaction with dipeptidyl peptidase 4, the viral receptor (Appendix Figure 1). We validated the specificity of the assay for human diagnostics using serum samples from healthy blood donors, PCR-confirmed non–coronavirus-infected patients and non–MERS-CoV–infected patients (cohorts H1–H3) (Appendix Table 1). At a 1/20 dilution, none of the samples from non–MERS-CoV-infected humans showed a >50% reduction in signal (RBI50) (Figure, panel A), indicating a high specificity of the assay. MERS-CoV–specific RBI antibodies were detected in all the 90% plaque reduction neutralization assay (PRNT90)–positive serum samples of the PCR-confirmed MERS-CoV patients tested (Appendix Table 2, Figure 2). The percentage reduction in signal strongly correlated with neutralizing antibody titers (Figure, panel B). The RBI50 assay showed similar sensitivity to the PRNT90 assay.
Because the RBI assay is species-independent, we validated its ability to detect RBI antibodies in dromedaries. At a 1/20 dilution, none of the naive dromedary serum samples (10) reacted in the assay, whereas all samples from MERS-CoV–infected dromedaries (2) resulted in a >90% reduction in signal (Appendix Table 1, Figure 3, panel A). We detected RBI antibodies in the samples of vaccinated and experimentally infected dromedaries (Appendix Figure 3, panel B). Overall, the RBI50 was highly specific and showed comparable sensitivity to PRNT90 for detection of MERS-CoV–specific RBI (neutralizing) antibodies after infection and vaccination (Appendix Figure 3, panel C).
Apart from the RBD, the MERS-CoV S1 contains an α2,3 sialic acid–binding S1A domain (7). When this domain was multivalently presented on self-assembling lumazine synthase (LS) nanoparticles (S1A-Np), it was able to hemagglutinate human erythrocytes. To generate S1A-Np, we genetically fused the S1A domain to LS and expressed the particles in HEK-293S cells (Appendix Figure 4, panel A). By using S1A-Np, we developed a hemagglutination inhibition (HI) assay to detect antibodies capable of blocking virus interaction with sialic acids (Appendix Figure 4, panel B). To set up the assay using turkey RBCs, we tested the ability of S1A-Np to agglutinate turkey erythrocytes by using empty (LS)-Np and S1B-Np as negative controls. Although neither the lumazine synthase–Np nor the S1B-Np showed any hemagglutination at any temperature tested, the S1A-Np induced hemagglutination at 4°C; we also noted hemagglutination when using dromedary erythrocytes (Figure, panel C; Appendix Figure 4, panel C). Although antibodies against the S1 and S1A domain inhibited hemagglutination showing high HI titers, S1B antibodies were negative for HI (Figure, panel D).
Next, we used the same cohort of serum samples for validating the RBI assay. None of the samples from healthy blood donors, PCR-confirmed non–coronavirus-infected and non–MERS-CoV–infected patients (cohorts H1–H3) showed any HI at the 1/20 dilution (Figure, panel E). HI antibodies were detected in the samples of all severely infected MERS-CoV patients and that of 1 mildly infected MERS-CoV patient (Figure, panel E; Appendix Figure 5); only 2 of the mildly infected MERS-CoV patients were PRNT90-positive (Appendix Table 2). Serum HI titers correlated strongly with neutralizing antibody titers detected by a whole virus neutralization assay (PRNT90); nonetheless, the PRNT90 assay was more sensitive (Figure, panel F). Similarly, only serum samples from MERS-CoV–infected dromedaries were HI-positive (10/13), whereas none of the naive dromedary camel serum samples showed any HI (Appendix Figure 6, panel A). HI antibodies were detected in serum samples of vaccinated dromedaries after booster immunization (Appendix Figure 6, panel B). Overall, although less sensitive, the antibody titers detected by the HI assay correlated strongly with the neutralizing antibody titers detected by PRNT90 assay (Appendix Figure 6, panel C).
The RBI and HI assays we developed are easy to operate and standardize and can detect functional antibodies against 2 MERS-CoV S1 domains responsible for virus entry (RBD) and attachment (S1A). Both assays are protein-based and can be carried out in a 96-well plate format, therefore providing BSL-1 high-throughput platforms. The assays can be used as confirmatory assays for human and dromedary MERS-CoV diagnostics and for antibody and vaccine evaluation.
Ms. Okba is a PhD candidate in the Viroscience Department at Erasmus Medical Center. Her research interests include the development of diagnostic and intervention strategies for emerging viruses.
Acknowledgment
This work was supported by the Zoonoses Anticipation and Preparedness Initiative (ZAPI project; Innovative Medicines Initiative grant agreement no. 115760), with the assistance and financial support of Innovative Medicines Initiative and the European Commission, in-kind contributions from European Federation of Pharmaceutical Industries and Associations partners. Proteins used for the assays described in this study can be provided by European Virus Archive–global. Requests should be submitted on the European Virus Archive–global website (http://www.european-virus-archive.com).
References
- World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV) [cited 2019 May 10].
- Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–5.
- Corman VM, Müller MA, Costabel U, Timm J, Binger T, Meyer B, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334.
- Reusken C, Mou H, Godeke GJ, van der Hoek L, Meyer B, Müller MA, et al. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveill. 2013;18:20441.
- Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagato O, et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574.
- Trivedi S, Miao C, Al-Abdallat MM, Haddadin A, Alqasrawi S, Iblan I, et al. Inclusion of MERS-spike protein ELISA in algorithm to determine serologic evidence of MERS-CoV infection. J Med Virol. 2018;90:367–71.
- Li W, Hulswit RJG, Widjaja I, Raj VS, McBride R, Peng W, et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A. 2017;114:E8508–17.
- Mou H, Raj VS, van Kuppeveld FJ, Rottier PJ, Haagmans BL, Bosch BJ. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol. 2013;87:9379–83.
- Widjaja I, Wang C, van Haperen R, Gutiérrez-Álvarez J, van Dieren B, Okba NMA, et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg Microbes Infect. 2019;8:516–30.
- Haagmans BL, van den Brand JM, Raj VS, Volz A, Wohlsein P, Smits SL, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;351:77–81.
Figure
Cite This ArticleOriginal Publication Date: 3/9/2020





















.png)












No hay comentarios:
Publicar un comentario