Finding Pediatric Cancer Genomic Data through PGDI
September 5, 2018, by Pamela Birriel, Ph.D. & Daniela S. Gerhard, Ph.D.
The Pediatric Genomic Data Inventory, or PGDI, was designed to list pediatric cancer genomic data available to researchers. The open-access inventory, launched in 2018 by the Office of Cancer Genomics (OCG), details the types of cancer studied, molecular characterization data available, and points of contact for each known project.
A Great Need for Pediatric Cancer Data
Childhood cancers constitute a diverse group of malignancies that are diagnosed in patients ranging in age from newborns to young adults. In 2017, an estimated 15,270 children and adolescents under the age of 20 were diagnosed with cancer in the United States.1Although cancer is the leading cause of death from disease in children,2 generating genomic data from their tumors remains a significant hurdle. Furthermore, many pediatric cancers are proving to have a genetically distinct component from their adult counterparts,3 demonstrating the need for childhood-specific genomic studies and therapeutic strategies.
OCG is committed to advancing the understanding of pediatric oncology and has helped generate many datasets through large-scale molecular characterization projects, such as the National Cancer Institute’s Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative. To expose information about the data, as well as other projects, OCG developed PGDI. The aim is to incorporate data descriptions from all pediatric genomic projects that are available for access by investigators.
OCG is committed to advancing the understanding of pediatric oncology and has helped generate many datasets through large-scale molecular characterization projects, such as the National Cancer Institute’s Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative. To expose information about the data, as well as other projects, OCG developed PGDI. The aim is to incorporate data descriptions from all pediatric genomic projects that are available for access by investigators.
Connecting Researchers with Data Through PGDI
Currently, PGDI includes 40 pediatric cancer sequencing cohorts or projects from the TARGETinitiative and the St. Jude–Washington University Pediatric Cancer Genome Project (PCGP). Projects comprise 2,524 enrolled cases across ~19 cancer types that span throughout the United States, Brazil, Canada, and Japan. However, we recognize that there are many more datasets out there and encourage other investigators to submit their project(s) to the PGDI list. A single, easy to use list, will enable researchers to find potential collaborations which in turn would increase our understanding of the causes of childhood cancers and allow development of improved therapies for sick children.
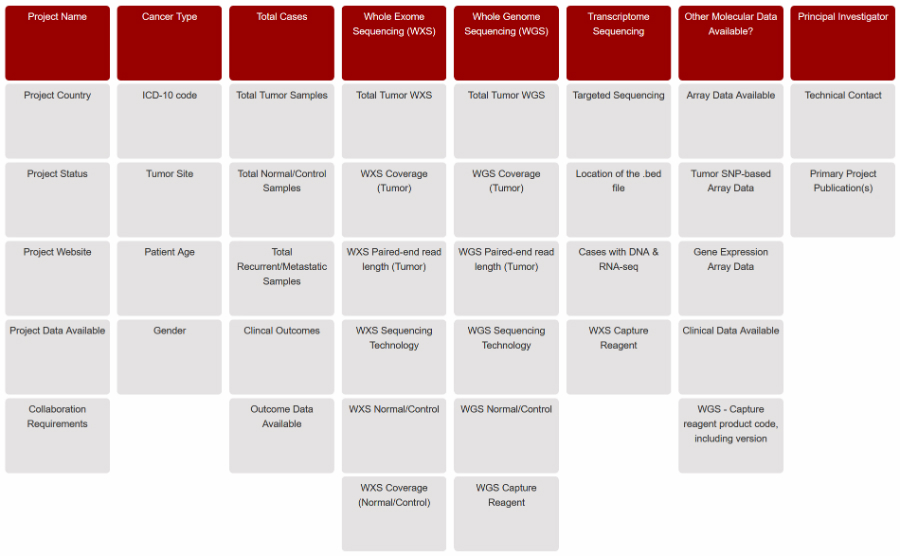
The PGDI Display allows users to view, sort, and search through the inventory. Individuals can view the default display categories, select up to 10 categories among cancer types and data (see figure) to create a custom view, and download the complete tab-delimited inventory.
The PGDI Display allows users to view, sort, and search through the inventory. Individuals can view the default display categories, select up to 10 categories among cancer types and data (see figure) to create a custom view, and download the complete tab-delimited inventory.
Community Engagement for a Public Resource
The potential for PGDI to further research and make an impact for pediatric patients depends on participation from the research community. Researchers are urged to participate and submit information about their pediatric molecular characterization cohorts or projectsby becoming a PGDI Data Contributor.
For more details about contributing to PGDI, read the OCG e-Newsletter article. For further help accessing and submitting data, contact OCG.
For more details about contributing to PGDI, read the OCG e-Newsletter article. For further help accessing and submitting data, contact OCG.
References
- National Cancer Institute. Cancer in Children and Adolescents. The website of the National Cancer Institute. https://www.cancer.gov/types/childhood-cancers/child-adolescent-cancers-fact-sheet. Accessed August 2018.
- National Cancer Institute. Childhood Cancers. The website of the National Cancer Institute. https://www.cancer.gov/types/childhood-cancers. Accessed August 2018.
- Ma X, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018 Mar 15;555(7696):371-376. doi:10.1038/nature25795



































No hay comentarios:
Publicar un comentario