
Lab Chat: Teaching machines to peek inside cells
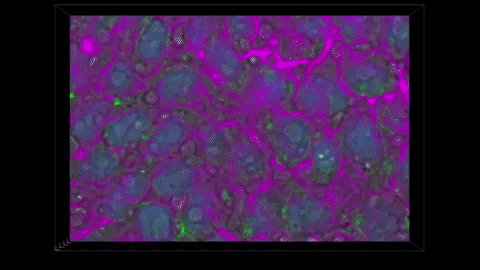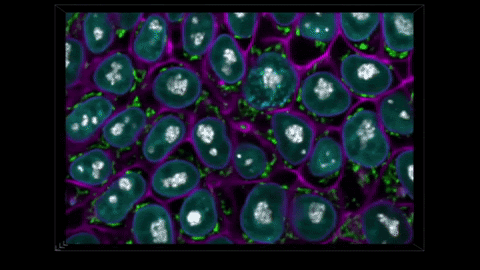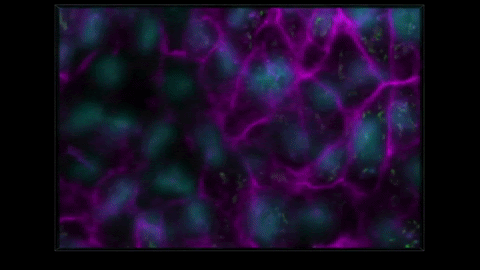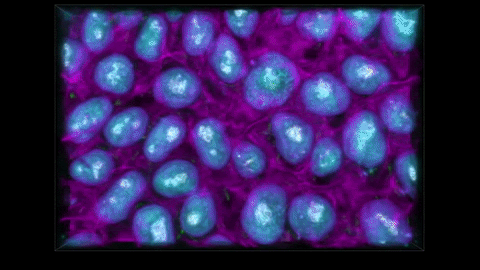
THE METHOD CAPTURES NUCLEAR ENVELOPES IN PURPLE, MITOCHONDRIA IN GREEN, AND CELL MEMBRANES IN PINK. (ALLEN INSTITUTE)
Scientists have trained computers to peer into living cells and detail their structures, which could reveal new clues about the state of healthy and diseased cells. Here’s what Molly Maleckar of the Allen Institute told me about the work, published in Nature Methods.
What problem did you set out to tackle?
One of the grand challenges in modern biology is to understand how the components inside of a cell work together. We have these bright-field microscopes like those you had in high school that produce black and white images relatively cheaply. There’s a lot of information in those images, but it looks jumbled and is hard for the human eye to see. And we have a way to image cells by adding fluorescent tags that let us see more, but cells can be damaged by that.
How did you try to solve that?
We have these pairs of images, one grayscale, one glow-in-the-dark. We trained a computer model to learn the relationship between the two. The idea is that you can take a grayscale image, feed it to the model, and the model will predict the fluorescent structure. You can do that for the mitochondria, the cell membrane, the nucleus, until you get an integrated look at a cell.
We have these pairs of images, one grayscale, one glow-in-the-dark. We trained a computer model to learn the relationship between the two. The idea is that you can take a grayscale image, feed it to the model, and the model will predict the fluorescent structure. You can do that for the mitochondria, the cell membrane, the nucleus, until you get an integrated look at a cell.


































No hay comentarios:
Publicar un comentario