
Volume 24, Number 2—February 2018
Dispatch
Emergomyces africanus in Soil, South Africa
On This Page
Ilan S. Schwartz , Barbra Lerm, J. Claire Hoving, Chris Kenyon, William G. Horsnell, W. Joan Basson, Patricia Otieno-Odhiambo, Nelesh P. Govender, Robert Colebunders, and Alfred Botha
, Barbra Lerm, J. Claire Hoving, Chris Kenyon, William G. Horsnell, W. Joan Basson, Patricia Otieno-Odhiambo, Nelesh P. Govender, Robert Colebunders, and Alfred Botha
Abstract
We detected Emergomyces africanus, a thermally dimorphic fungus that causes an HIV-associated systemic mycosis, by PCR in 18 (30%) of 60 soil samples from a wide range of habitats in South Africa. Direct and indirect culture techniques were unsuccessful. Experimental intraperitoneal inoculation of conidia induced murine disease.
The newly described thermally dimorphic fungal genus Emergomyces comprises human pathogens that cause systemic mycoses in immunocompromised persons globally (1). Among these fungi, Emergomyces africanus(formerly Emmonsia sp. [2]) is the species responsible for the most human disease. HIV-associated emergomycosis is the most common endemic mycosis in South Africa and is associated with a high case-fatality ratio (3,4).
Although an environmental reservoir for Es. africanus has not been established, soil is presumed to harbor the mycelial phase (2). We tested soils in South Africa for Es. africanus by using molecular- and culture-based methods.
We collected 60 soil samples from various soil habitats around South Africa by convenience sampling; 82% percent of samples came from the Western Cape Province, with the remaining samples from Gauteng (7%), Eastern Cape (7%), KwaZulu-Natal (2%), and Northern Cape (2%) provinces. For each sample, we used sterile, plastic tubes to collect ≈100 mL of topsoil.
We extracted DNA from soil by using the ZR Soil Microbe DNA Miniprep Kit (Zymo Research, Irvine, CA, USA). DNA extraction was successful for 56 soil samples (93%). We subjected extracted genomic DNA (gDNA) to a nested PCR. To amplify the internal transcribed spacer (ITS) region of the ribosomal RNA, we used the universal primers ITS1 and ITS4 in the first reaction (5). We used an Applied Biosystems 2720 Thermal Cycler (Foster City, CA, USA); thermocycling conditions consisted of 95°C for 5 min, 30 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 45 s, and 72°C for 7 min. We subjected PCR products to amplification by using Es. africanus–specific primers (forward, 5′-CCTGGTTTGGGGAGAGGGGT-3′; reverse, 5′-CCGGGGGAGCTCTTGGCTCT-3′), followed by electrophoresis on a 2% agarose gel. We performed amplification as described, except with an annealing temperature of 57°C. PCR mixtures consisted of 10 µL 2× KAPA Taq ReadyMix (KAPA Biosystems, Wilmington, MA, USA); 1 µL of each primer (10 µmol/L; Inqaba Biotechnical Industries, Pretoria, South Africa); and 1 µL of extracted gDNA or ITS PCR product, in a final reaction volume of 20 µL. We sequenced amplified products and compared them using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The PCR could detect as few as 102–104 conidia/10 g of soil (Technical Appendix[PDF - 405 KB - 8 pages]).
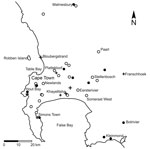
We plotted results of molecular testing and residential postal codes of persons with confirmed infections (Figure 1). We detected Es. africanus DNA in 18 (32%) of 56 soil samples representing all types of soil habitats tested (Table).
We used soil dilution plates prepared with Sabouraud agar (40 g/L glucose [Merck, Darmstadt, Germany], 10 g/L peptone [Merck], and 15 g/L agar) supplemented with 0.2 g/L chloramphenicol (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) to culture Es. africanus from 4 randomly selected soil samples. We incubated the resulting spread plates at 26°C, inspecting plates daily for 1 week and then twice weekly for an additional 3 weeks. All culture plates were rapidly overgrown by filamentous fungi other than Es. africanus.
To overcome rapid contamination, we used indirect culture methods. First, we used the flotation method adapted from Larsh et al. (7) to separate the conidia from other particles in the soil (Technical Appendix[PDF - 405 KB - 8 pages]). We plated the resulting soil suspensions on Sabouraud agar and brain heart infusion (BHI) plates and incubated them at 26°C, conducting daily examinations for fungal colonies resembling Es. africanus (1). This preparation also resulted in rapid contamination of all plates.
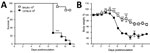
Figure 2. Infection of mice with Emergomyces africanus. In a proof-of-principle study, C57BL/6 and BALBc mice were inoculated intraperitoneally with 106 conidia (Es. africanus CAB 2141, a clinical isolate) in saline. Mice were...
Thereafter, we passaged soil suspensions through mice to screen out nonpathogenic soil organisms (8; Technical Appendix[PDF - 405 KB - 8 pages]). Animal studies were approved by the University of Cape Town’s Animal Ethics Committee (protocol 016–002). We created soil suspensions by using the flotation method and sampling from the bottom third of the column; penicillin G (1,000 IU/mL) and gentamicin (0.1 mg/mL) were included in the solution. We inoculated 1 mL of soil suspension intraperitoneally into each of 4 BALB/c or C57BL6 mice. We euthanized the mice after 2 weeks and plated livers, spleens, or both onto Sabouraud agar plates with and without chloramphenicol, which we then incubated at 30°C and 35°C –37°C. We inspected plates as described previously. Pilot studies demonstrated that this method could detect as few as 102 conidia in 10 g of soil (Technical Appendix[PDF - 405 KB - 8 pages]). Notably, in a pilot study in which BALB/c and C57BL/6 mice were challenged with graded doses of Es. africanus conidia, genetic background of mice influenced host susceptibility to the organism; C57BL/6 mice were more sensitive to infection and had significantly higher mortality and weight loss in response to the high dose of 106 conidia compared with BALB/c mice (Figure 2).
We screened 26 soil samples for the presence of Es. africanus by using mouse passage. These samples included all 18 soil samples in which Es. africanuswas detected by nested PCR, as well 8 soil samples that were PCR-negative. None of these samples, however, led to the isolation of Es. africanus through mouse passage.
Es. africanus is a newly described dimorphic fungal pathogen and causes an important HIV-associated systemic mycosis in South Africa (9). Many aspects of this organism remain unknown, including its ecologic niche. Our findings demonstrate that Es. africanus is present in a high proportion of soil samples collected from a range of habitats in South Africa, suggesting that soil might be a natural reservoir for this pathogen.
The isolation of pathogenic fungi from soil is challenging. Soil naturally contains a vast array of bacteria, viruses, fungi, and protozoa, all of which can interfere with or contaminate culturing the organism of interest (8). Since 1932, when Stewart and Meyer first cultured Coccidioides immitis from soil (10), flotation and animal passage has been the most robust method to isolate pathogenic fungi from soil. However, animal passage is laborious and expensive, can take months of turnaround time, requires special animal facilities, and results in discomfort and loss of life to laboratory animals, necessitating stringent ethics review (11).
Molecular detection is a valuable tool for establishing the presence of genetic material in environmental samples (11). In addition to high sensitivity, molecular detection has the advantages of being easy to apply, inexpensive, and rapid, and it can be performed in most laboratories. Alternatively, molecular detection lacks specificity because it cannot determine the viability (and hence infectivity) of the detected target (11). In our study, mouse passage of soil samples shown by nested PCR to contain Es. africanus genetic material did not result in the isolation of this fungus.
We have demonstrated that experimental infection with Es. africanus can produce pathology in mice. Moreover, susceptibility to disease appears to be mouse strain–dependent, with C57BL/6 mice being more susceptible than BALB/c mice.
This study has some limitations. The number of samples, and especially those from outside Western Cape Province, was relatively small, limiting inferences about the geographic range of Es. africanus in the environment. Moreover, our method of convenience sampling is prone to sampling bias. Nonetheless, this study is instructive for future ecologic studies, which should use random sampling to refine knowledge of the ecologic niche of this fungus.
In conclusion, this study demonstrates that Es. africanus can be frequently detected in a wide range of soils in South Africa. Moreover, our findings support the hypothesis that soil serves as a reservoir for this pathogen.
Dr. Schwartz is an infectious diseases physician and researcher with interests in emerging fungal infections, immunocompromised hosts, and global health. This work comprises part of a doctoral thesis at the Global Health Institute, Faculty of Medical Sciences, University of Antwerp.
Acknowledgments
The authors acknowledge the contributions of Kim Hoek and Mae Newton-Foot for help with primer design, Andrew Whitelaw for laboratory support, Kim Tutt for guidance on the animal inoculation protocol, Marco Botha for creating the map used in Figure 1, and Kristine Schwartz and the students of Alfred Botha’s biology class for help with sample collection.
This study was funded by the Fonds Wetenschappelijk Onderzoek–Vlaanderen (G.0514.14N). I.S.S. was supported by an Association of Medical Microbiologists and Infectious Diseases Physicians (AMMI) Canada – Astellas Post-Residency Fellowship (2015) and a grant from the R. Samuel McLaughlin–Manitoba Medical Services Foundation and University of Manitoba Dean’s Fellowship Fund (2016). J.C.H. is funded by the South African National Research Foundation and Carnegie Corporation.
References
- Dukik K, Muñoz JF, Jiang Y, Feng P, Sigler L, Stielow JB, et al. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae(Onygenales). Mycoses. 2017;60:296–309. DOIPubMed
- Schwartz IS, Kenyon C, Feng P, Govender NP, Dukik K, Sigler L, et al. 50 years of Emmonsia disease in humans: the dramatic emergence of a cluster of novel fungal pathogens. PLoS Pathog. 2015;11:e1005198. DOIPubMed
- Maphanga TG, Britz E, Zulu TG, Mpembe RS, Naicker SD, Schwartz IS, et al. In vitro antifungal susceptibility of the yeast- and mould-phases of the dimorphic fungal pathogen, Emergomyces africanus (formerly Emmonsia species), from HIV-infected South African patients. J Clin Microbiol. 2017;55:1812–20. DOIPubMed
- Schwartz IS, Govender NP, Corcoran C, Dlamini S, Prozesky H, Burton R, et al. Clinical characteristics, diagnosis, management and outcomes of disseminated emmonsiosis: a retrospective case series. Clin Infect Dis. 2015;61:1004–12. DOIPubMed
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. p. 315–22.
- Schwartz IS, Kenyon C, Lehloenya R, Claasens S, Spengane Z, Prozesky H, et al. AIDS-related endemic mycoses in Western Cape, South Africa, and clinical mimics: a cross-sectional study of adults with advanced HIV and recent-onset, widespread skin lesions. Open Forum Infect Dis. 2017;4:ofx186. DOIPubMed
- Larsh HW, Hinton A, Furcolow ML. Laboratory studies of Histoplasma capsulatum. III. Efficiency of the flotation method in isolation of Histoplasma capsulatum from soil. J Lab Clin Med. 1953;41:478–85.PubMed
- Emmons CW. The isolation from soil of fungi which cause disease in man. Trans N Y Acad Sci. 1951;14(1 Series II):51–4. DOIPubMed
- Kenyon C, Bonorchis K, Corcoran C, Meintjes G, Locketz M, Lehloenya R, et al. A dimorphic fungus causing disseminated infection in South Africa. N Engl J Med. 2013;369:1416–24. DOIPubMed
- Stewart R, Meyer K. Isolation of Coccidioides immitis (Stiles) from the soil. Proc Soc Exp Biol Med. 1932;29:937–8. DOI
- Li Z, Chaturvedi V. The powers and perils of PCR in the search for the natural reservoirs of Coccidioides species. Mycopathologia. 2017;182:435–8. DOIPubMed






















.png)











No hay comentarios:
Publicar un comentario