
DNA-Encoded Movie Points Way to ‘Molecular Recorder’
Credit: Seth Shipman, Harvard Medical School, Boston
There’s a reason why our cells store all of their genetic information as DNA. This remarkable molecule is unsurpassed for storing lots of data in an exceedingly small space. In fact, some have speculated that, if encoded in DNA, all of the data ever generated by humans could fit in a room about the size of a two-car garage and, if that room happens to be climate controlled, the data would remain intact for hundreds of thousands of years! [1]
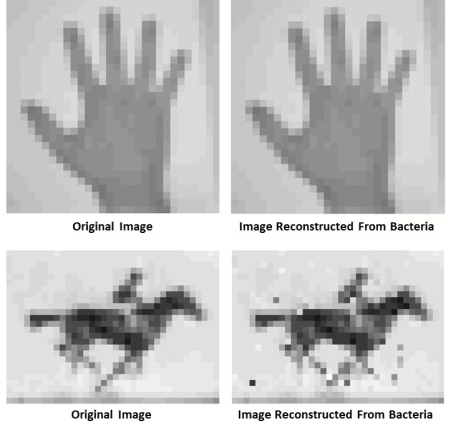
Scientists have already explored whether synthetic DNA molecules on a chip might prove useful for archiving vast amounts of digital information. Now, an NIH-funded team of researchers is taking DNA’s information storage capabilities in another intriguing direction. They’ve devised their own code to record information not on a DNA chip, but in the DNA of living cells. Already, the team has used bacterial cells to store the data needed to outline the shape of a human hand, as well the data necessary to reproduce five frames from a famous vintage film of a horse galloping (see above).
But the researchers’ ultimate goal isn’t to make drawings or movies. They envision one day using DNA as a type of “molecular recorder” that will continuously monitor events taking place within a cell, providing potentially unprecedented looks at how cells function in both health and disease.
The Harvard Medical School team, led by Seth Shipman and George Church, built their molecular recorder using the CRISPR/Cas complex, much touted on this blog as a gene-editing tool. It relies heavily on the Cas9 enzyme, often likened to a pair of molecular scissors for its ability to target specific DNA sequences and cut them. But the CRISPR complex, discovered in the adaptive immune system of bacteria, includes other parts too.
In the new work, Shipman and Church relied on two of those other parts: Cas1 and Cas2. These enzymes allow bacteria to capture nucleotide sequences snipped from invading viruses and store them in their genomes for future reference. Church’s group had earlier shown they could direct the CRISPR system to capture and store lab-made synthetic DNA sequences. In the study recently reported in the journal Nature, they wanted to see if they could use those CRISPR/Cas parts to record information inside living cells [2].
The first step was to devise a coding scheme for converting images into DNA nucleotides. In their first scheme, they simply assigned each of the four DNA bases (A, C, G and T) to a pixel color. In the second, they devised a more flexible system including 21 different pixel colors. Each color was specified by a different combination of three nucleotide bases.
In the first demonstration, the researchers converted the outline of a hand, pixel by pixel, using each of their DNA codes. The plan was then to introduce their DNA code into the common bacterium Escherichia coli. In E. coli, the CRISPR system takes up DNA from invading viruses in 33-base units called spacers. So, the researchers chopped their encoded image into a series of sequences, fitting them into thousands of spacers and delivering them sequentially to a population of E. coli bacteria in the lab.
The researchers left the cells to grow and divide overnight. The next day, they sequenced the DNA and tried piecing the image back together. The researchers found they could reconstruct the images with up to 96 percent accuracy.
The next challenge was to see if they could use their code and molecular recorder to capture and later recall multiple images over time—even to create movies. They selected five frames of the mare ‘Annie G.’ galloping in the 1870s film Human and Animal Locomotion. As Shipman noted, it was one of the first moving pictures ever captured on film, using technology that, like CRISPR today, was considered revolutionary back then. The researchers found they could reconstruct each movie frame and the order of those frames with better than 90 percent accuracy.
The findings suggest the molecular recorder could one day be used to capture other types of information in a cell over time. For instance, Shipman says he hopes ultimately to insert the molecular recorder into brain cells, where it could be used to log changes in gene activity continuously for an unprecedented look at brain development.
Another intriguing possibility is to use this approach in bacteria that naturally inhabit our bodies or the natural environment. For instance, the bacteria could be programmed to function as living biosensors, recording, for example, the presence of heavy metals or other contaminants over time.
Now that the team has shown it can use the CRISPR system to capture, store, and retrieve data that’s been fed into bacterial cells, the next challenge is to see if the system can capture biological data that’s new and unknown. If so, their results promise to open an entirely new window for discovery.
References:
[1] DNA could store all of the world’s data in one room. Science. 2 March 2017.
[2] CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Shipman SL, Nivala J, Macklis JD, Church GM. Nature. 2017 July 12. [Epub ahead of publication]
Links:
Popular Genome Editing Tool Gets Its Close-Up (NIH Director’s Blog)
Church Lab (Harvard Medical School)
NIH Support: National Institute of Mental Health; National Institute of Neurological Disorders and Stroke; National Human Genome Research Institute






















.png)











No hay comentarios:
Publicar un comentario